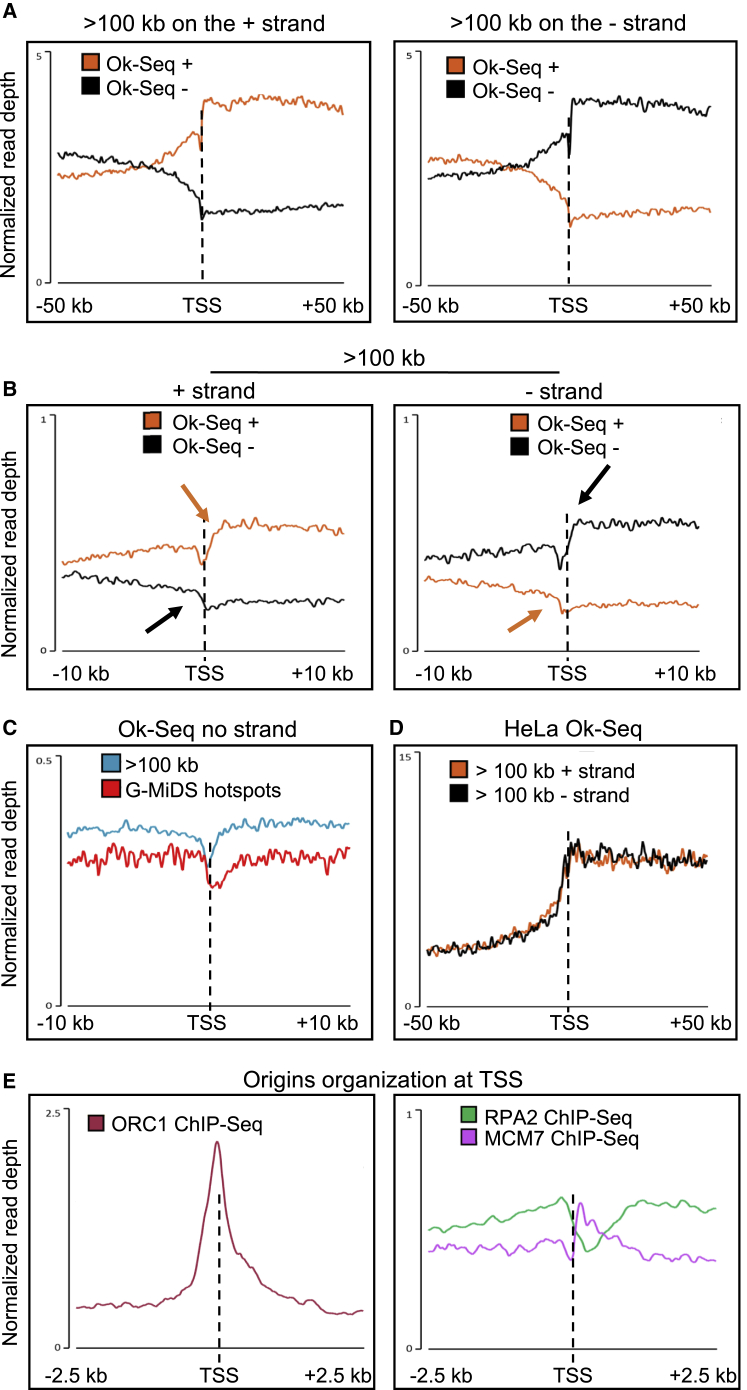
Figure 5.
Uncoupling of replication forks efficiency at origins of replication near TSSs
(A) Average metagene profile for the denotated strand of strand-specific Ok-seq from Chen et al. (2019) TSSs ±50 kb of transcribed genes >100 kb in BJ-hTERT cells.
(B) As for (A) but for TSSs ±10 kb, with orange and black arrows indicating the start positions of the Okazaki fragments on “+” or “−” strands.
(C) Average metagene profile for Ok-seq from Chen et al. (2019) TSSs ±10 kb of transcribed genes >100 kb or G-MiDS hotspot genes in BJ-hTERT without strand specificity.
(D) Average metagene profile for Ok-seq transcribed/not-transcribed strand from Petryk et al. (2016) TSSs ± 50 kb of transcribed genes >100 kb in HeLa cells on + or − strands.
(E) Average metagene profile of MCM7 (Sugimoto et al., 2018), RPA2 (Zhang et al., 2017), and ORC1 (Dellino et al., 2013) ChIP-seq in HeLa cells at TSSs of transcribed genes >100 kb.