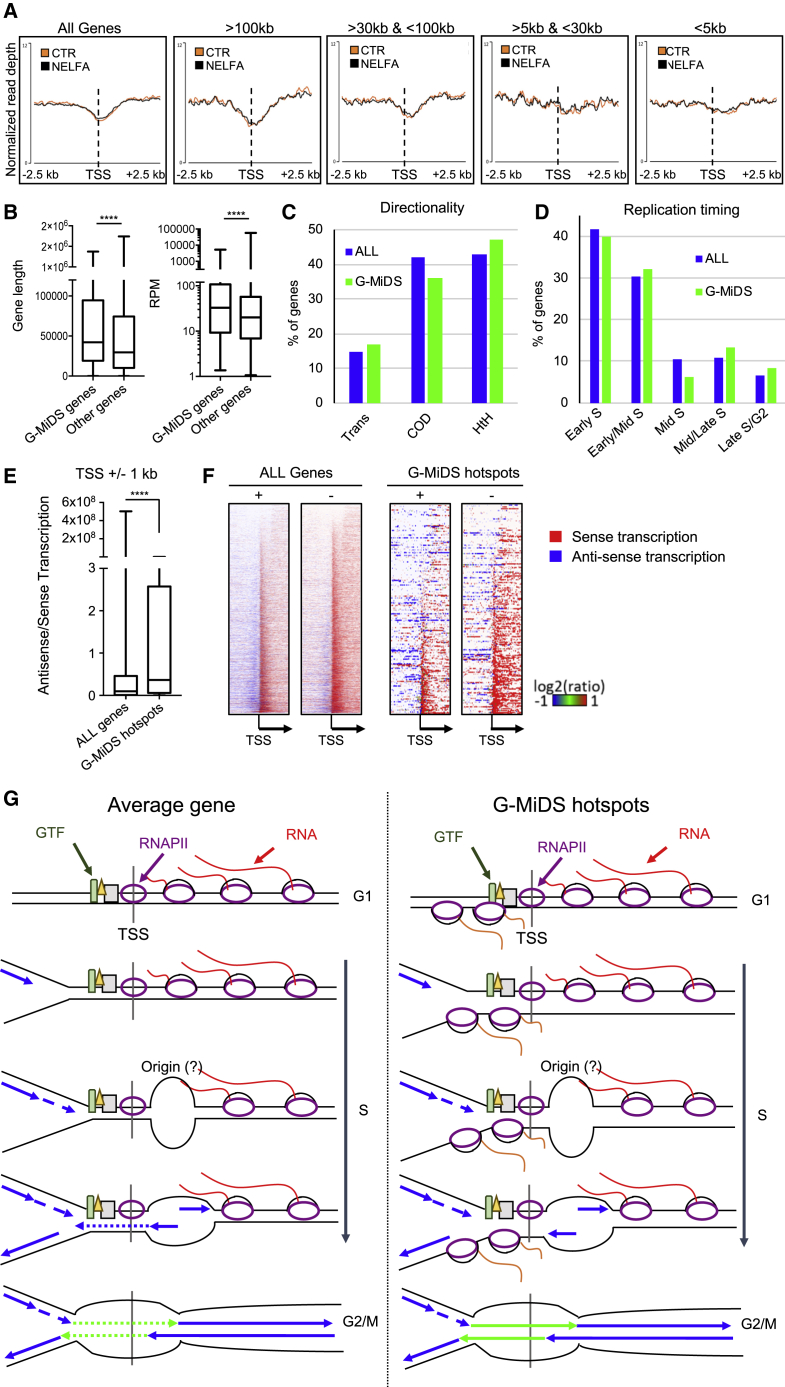
Figure 7.
G-MiDS-hotspot-gene-specific features
(A) As in Figures 3A and 3B for all transcribed genes and transcripts separated by gene length in cells transfected with the indicated siRNA.
(B) Gene length and transcription levels at the late-S/G2 time point for G-MiDS hotspot genes toward all other transcribed genes.
(C) Directionality analysis of G-MiDS hotspot genes compared with directionality of all transcribed genes.
(D) Replication timing of G-MiDS hotspot genes compared with all transcribed genes.
(E) Quantification of the ratio between antisense and sense transcription at TSSs ±1 kb of all transcribed genes and the 449 G-MiDS hotspots.
(F) Heatmap analysis of the levels of antisense and sense transcription at TSSs ±2.5 kb for all the transcribed genes and G-MiDS hotspots on the + and − strands.
(G) Model describing how the TSS is occupied by RNAPII and general transcription factors (GTF) throughout the cell cycle, with the RNAPII moving along genes. When DNA replication approaches the TSS during S phase, it may encounter GTF/RNAPII, skipping the TSS and restarting downstream of it. This may be mediated by the activation of origins of replication near TSSs. Later during S phase, DNA replication may fill the resulting gaps. However, in cases of genes with high steady expression levels of sense and TSS-associated antisense transcription, completion of the duplication of TSSs will occur in G2/M, when RNAPII and GTF are removed from TSSs. Box and whisker plots with the line at the median; Mann-Whitney t test; ∗∗∗∗p < 0.0001.