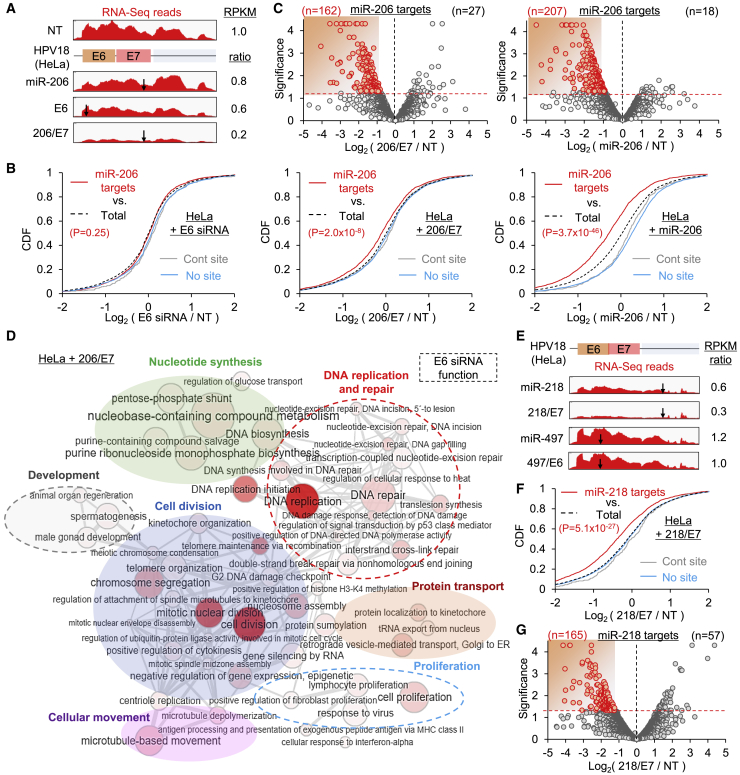
Figure 4.
Transcriptome-wide validation of the anticancer mi/siRNAs 206/E7 and 218/E7
(A) RNA-seq reads from NT-, miR-206-, E6-, and 206/E7-transfected HeLa cells, mapped on the HeLa-specific E6/E7 transcript. E6, E6 siRNA; reads per kilobase of transcript per million mapped reads (RPKM) ratio are shown relative to NT; arrows denote cognate target sites. (B) CDF analyses of putative miR-206 targets that contain seed sites in 3ʹ UTRs, depending on their fold changes (log2 ratio) in E6 (left panel), 206/E7 (middle panel), and miR-206 (right panel), relative to NT transfection. RPKM values are from Cufflinks; p values are from KS tests, relative to total transcripts (Total); “Cont site” indicates transcripts with non-nucleation bulge sites in 3ʹ UTRs, previously used as negative control;13 “No site” indicates transcripts with no miR-206 seed site in 3ʹ UTRs. (C) Volcano plot analyses of the putative miR-206 targets for 206/E7 (left panel) and miR-206 transfection (right panel). Significance, −log10(p value); downregulated DEG (p < 0.05; Cuffdiff), highlighted in red. (D) GO analysis results of the downregulated DEG in 206/E7-transfected HeLa cells (red dots in Figure S3D), represented as networks of overrepresented biological process terms (EASE score < 0.2, DAVID; node size and color intensity inversely correlate with p value). Clusters of terms functionally enriched in E6 siRNA transfection (Figure S4B) are highlighted with dotted circles. Of note, the rest of the clustered networks were derived from miR-206 function (Figures S3E and S4C), although there were some overlaps in the function of regulating development. (E) RNA-seq reads on E6/E7 transcript as analyzed in (A) except for miR-218, 218/E7, miR-497, and 497/E6 expression. The RPKM ratio is relative to 497/E6. (F and G) CDF (F) and volcano plot (G) analyses as performed in (B) and (C) except for putative miR-218 targets in 218/E7-transfected HeLa cells. Red dots indicated downregulated DEGs. All p values are from two-sided KS tests, relative to total, unless otherwise indicated.