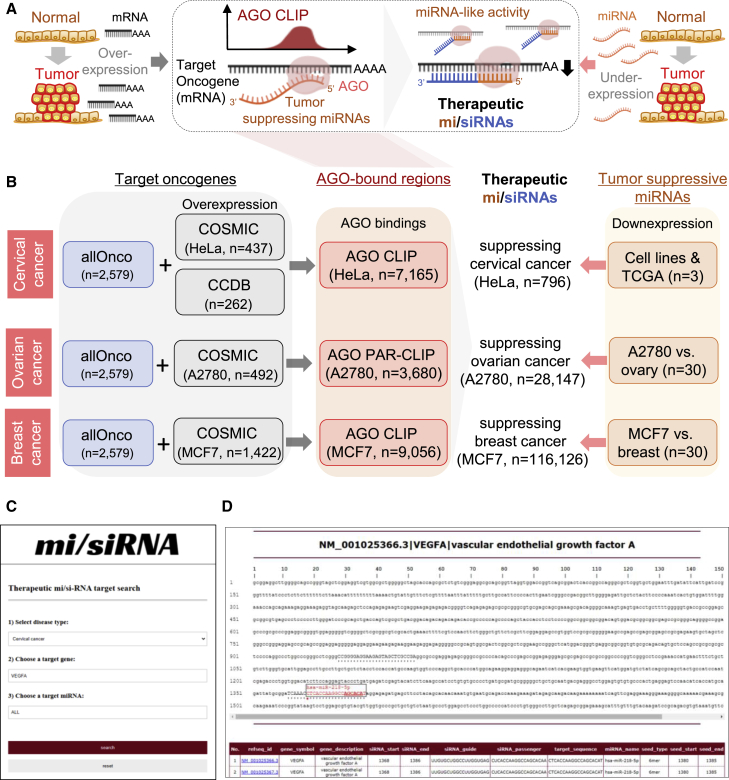
Figure 6.
Strategy of designing antitumor mi/siRNAs applied to cervical, ovarian, and breast cancers
(A) Overview of designing therapeutic mi/siRNAs based on AGO CLIP analysis for cancer treatment, conferring dual roles as siRNA-targeting oncogenes (overexpressed in tumor) and as tumor-suppressive miRNAs (underexpressed in tumor). (B) Antitumor mi/siRNAs delineated for cervical, ovarian, and breast cancers based on AGO CLIP results in HeLa cells,11 A2780,16 and MCF7.17,18 For selection of target oncogenes, a combination of allOnco (http://www.bushmanlab.org/links/genelists) and COSMIC databases was used. For cervical cancer, the list of amplified or overexpressed genes in the cervical cancer gene database (https://webs.iiitd.edu.in/raghava/ccdb/) was also used. Tumor-suppressive miRNAs were selected based on miRNA expression profiles derived from TCGA, the gene expression omnibus, and the microRNA (http://www.microrna.org) databases (details in Materials and methods). (C and D) The mi/siRNA web server (http://ago.korea.ac.kr/misiRNA) with searchable functionality for cervical, ovarian, and breast cancers (C) and an example of output results (D) are shown. Red asterisk indicates start position of the mi/siRNA target site; a black dot indicates AGO-bound regions. Of note, searches in the mi/siRNA web server could be started by defining a type of cancer (cervical, ovarian, or breast cancer), followed by autocompletion of the selected target oncogenes and tumor-suppressing miRNAs. Indication of AGO-bound regions (black dot, D) with detailed information from RefSeq and miRBase links could guide users to narrow down their choices.