Fig. 1.
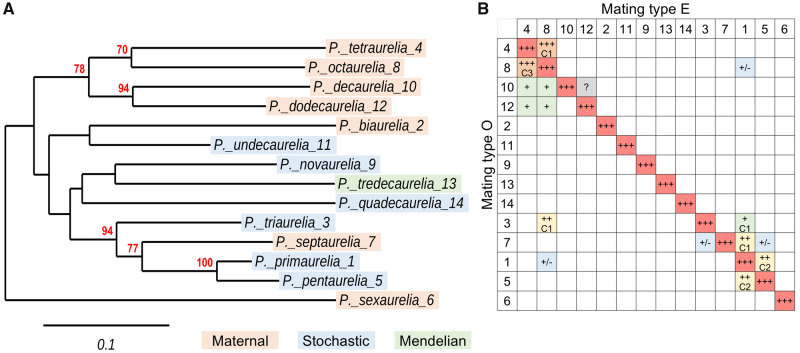
Phylogeny of the 14 classical Paramecium aurelia species and interspecies mating reactions. (A) COII phylogeny. The phylogenetic tree of the mitochondrial COII gene identifies groups of closely related species among the 14 “classical” species (also identified by the original “variety” numbers after binomial names). Bootstrap support values are shown only when ≥50%; these nodes are confirmed in the multi-gene tree of sequenced species shown in supplementary figure S1, Supplementary Material online. Together with the mtA gene tree in figure 2 and the mtB gene tree in supplementary file S1, Supplementary Material online the COII tree clarifies the correct phylogenetic position of Paramecium septaurelia, which was erroneous in many previous publications due to wrong species assignments (see Extended Data figs. 6–8 in Singh et al. 2014). The strains used are: Paramecium primaurelia AZ9-3, Paramecium biaurelia V1-4, Paramecium triaurelia 325, Paramecium tetraurelia 51, Paramecium pentaurelia 87, Paramecium sexaurelia AZ8-4, Paramecium septaurelia 227, Paramecium octaurelia 138, Paramecium novaurelia TE, Paramecium decaurelia 223, Paramecium undecaurelia 219, Paramecium dodecaurelia 274, Paramecium tredecaurelia 321, Paramecium quadecaurelia N1A. Color shadings indicate the mechanism of mating-type determination in each species: orange, maternal inheritance; blue, stochastic determination; green, Mendelian. (B) Mating reactions between species. The 14 species are designated by numbers, as in (A). The diagonal (red) represents intraspecies reactions between O and E clones, where both agglutination and pair formation show the best efficiency. The table reports the efficiency of all observed interspecies agglutination reactions, always between opposite mating types: +++ (orange), ∼90–95% of cells in clumps; ++ (yellow), ∼40% of cells in clumps; + (green), only a few % of cells in clumps; +/– (blue), brief and weak contacts one pair at a time; ? (gray), dubious. Whenever observed, the formation of conjugating pairs is also quantified: C3, ∼90% of clumped cells; C2, 40–50% of clumped cells; C1, a few % of clumped cells. The data are from Sonneborn (1975a).