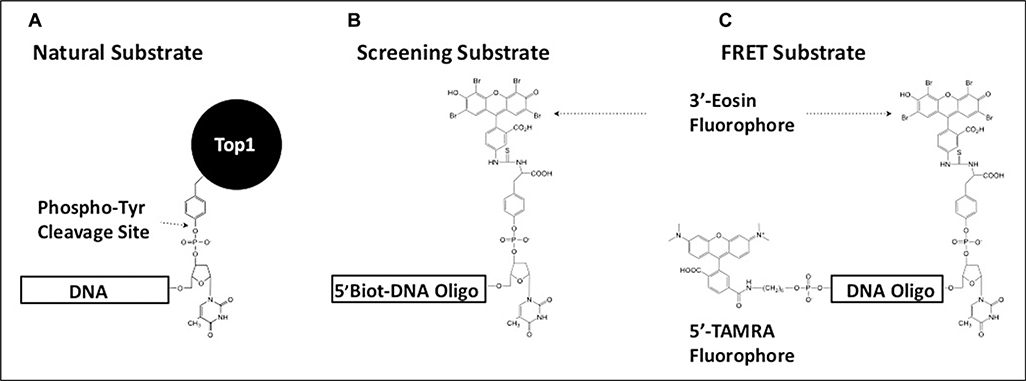
Figure 1.
Design of tyrosyl-DNA phosphodiesterase 1 (TDP1) substrates used in this study. (A) The natural cellular substrate of TDP1 was used as a model for the design of two fluorescent substrates. (B) The presence of a 5′-biotin moiety on the screening substrate allowed for immobilization on streptavidin-coated plates. This facilitated the inclusion of a postreaction wash step to remove cleaved 3′-eosin fluorescent product prior to measuring residual fluorescent signal from the remaining intact substrate. (C) By replacing the 5′-biotin moiety with a TAMRA fluorphore, a Förster resonant energy transfer substrate was designed for use during inhibitor characterization. TAMRA quenches the fluorescence emission signal in the intact substrate molecule. Cleavage of the substrate by TDP1 removes this interference, producing an increase in eosin emission signal that can be monitored continuously.