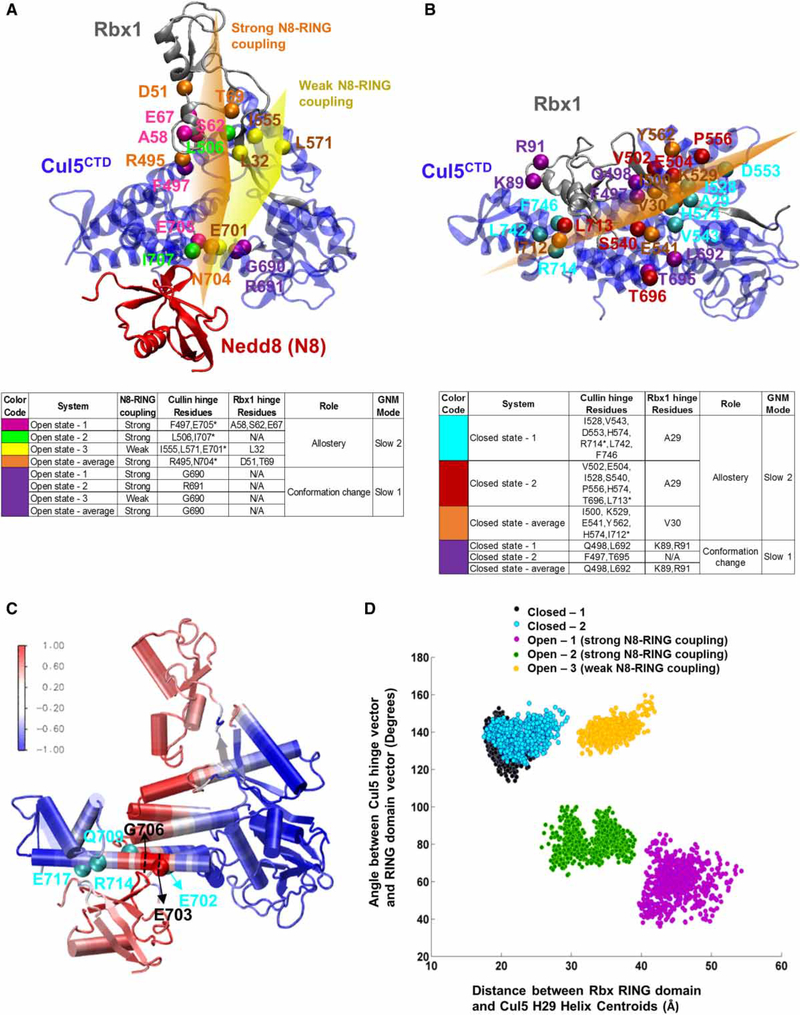
Figure 3. Hinge sites and coupled Cul regions on CRLCTD provide a dynamic measure for RING domain rotation.
Open-1 and closed-1 CRLCTD conformers are shown in (A and C) and (B), respectively, to illustrate the hinge sites. The open and closed state CRLCTD (Cul5CTD–Rbx1) hinge residues of the slowest mode of motion during Cul neddylation are shown in purple. Hinge residues coordinating the allosteric communication are colored differently for each conformer. Open state-average and closed state-average yield the average conformers for both states. Side tables are provided for guidelines. The Cul5 H29 hinge residues are marked with asterisk in the side tables. Approximate imaginary planes are drawn for illustration of hinge coordination. In (A), the orange and yellow hinge planes represent the hinge coordination for Nedd8–Cul5–Rbx1 allosteric communication when Nedd8–RING coupling is high and weak, respectively. In (B), the orange plane represents the hinge coordination for Cul5–Rbx1 allosteric communication in closed state conformers. (C) Open state CRLCTD conformer is colored according to the Cul5 flexible element showing highest correlation with Nedd8, Glu703–Gly706, to highlight the allosteric communication pathway from Nedd8 through CRLCTD. The normalized correlation values are in the range of [−1,1]. The Cul5 residues that H-bond with Nedd8 are shown in cyan. (D) Dynamic RING domain positions throughout each MD simulation are depicted; x-axis: distance between the RING domain (Cys42–Lys105) centroid and the Nedd8-binding Cul5 WHB domain’s H29 (Trp695–Met725) centroid, y-axis: angle between the state-specific hinge vector and the RING domain vector from Rbx1 Cys42 to the RING domain centroid.