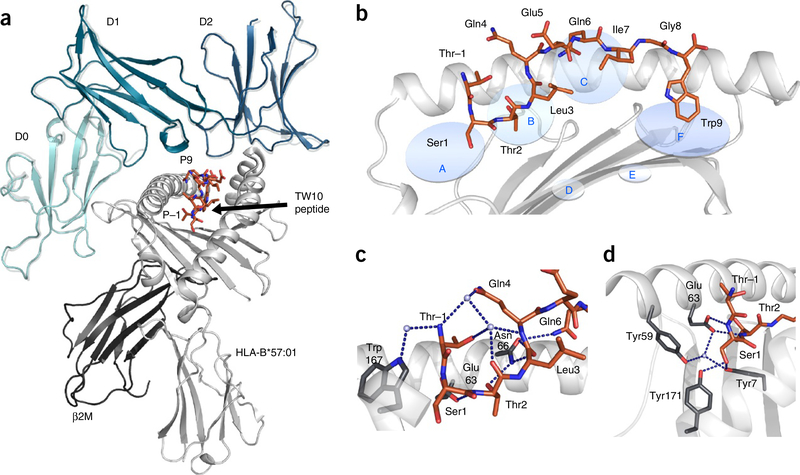
Figure 1.
HLA-B*57:01 in complex with the TW10 peptide. (a) The overall structure of KIR3DL1 (green) in complex with HLA-B*57:01–TW10 (orange). The structure of KIR3DL1 when bound to HLA B*57:01–LF9 is overlaid12 (gray) for comparison of the binding modes. (b) Cartoon representation of the crystal structure of HLA-B*57:01 (light gray) in complex with the TW10 peptide TSTLQEQIGW (orange) shown against the α1 helix of HLA and oriented N-C-terminal from left. Anchor pockets of the HLA are indicated along the binding groove at P1 (A) to PΩ (F). (c,d) Orientation of the protruding Thr residue of the peptide at P–1 (orange) and its interaction with residues of the HLA (dark gray sticks) (c), and the conserved hydrogen-bonding network (blue dashed lines) at the N-terminal end of HLA-B*57:01 (dark gray sticks) maintained to Ser at P1 (orange) (d). Hydrogen bonds are shown as blue dashed lines.