FIGURE 4.
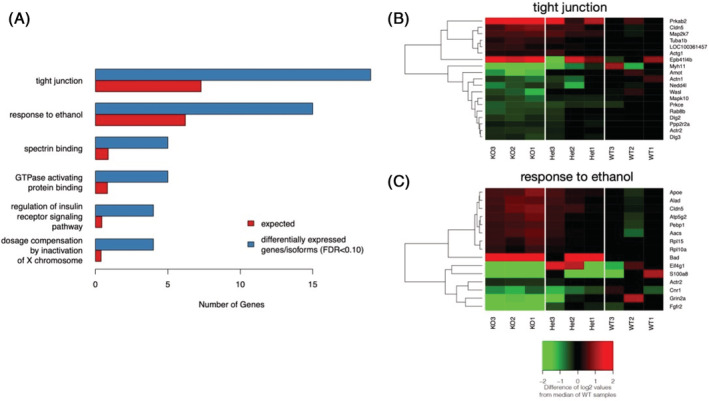
Functional enrichment of KEGG pathways and Gene Ontology (GO) terms in transcripts differentially expressed between Lrap knockout and wild type rat brains. Genes associated with isoforms and/or SOI that were differentially expressed (FDR < 0.10) were included. The background data set for enrichment include genes with at least one isoform and/or SOI that was expressed in brain and tested for differential expression. (A) Terms and pathways enriched for isoforms/SOI whose expression levels were altered by the genetic manipulation of Lrap. All pathways/terms included in the figure: (1) contained 3 or more differentially expressed isoform/SOI, (2) meet a significance threshold of an FDR <0.10, and (3) had a fold enrichment (observed number of differentially expressed genes divided by the number of differentially expressed genes expected by chance) of at least 2. Fifteen GO terms met all three criteria but only the top 5 (by p value) are included in the graphic for simplicity. Differentially expressed isoforms/SOI associated with the (B) Tight Junction KEGG pathway or associated with the (C) Response to Ethanol GO term. Expression values are represented in this heatmap as the difference in log2 transformed and library size adjusted read counts for a sample and the median of this transformed read count in the wild type rats
