Figure 2.
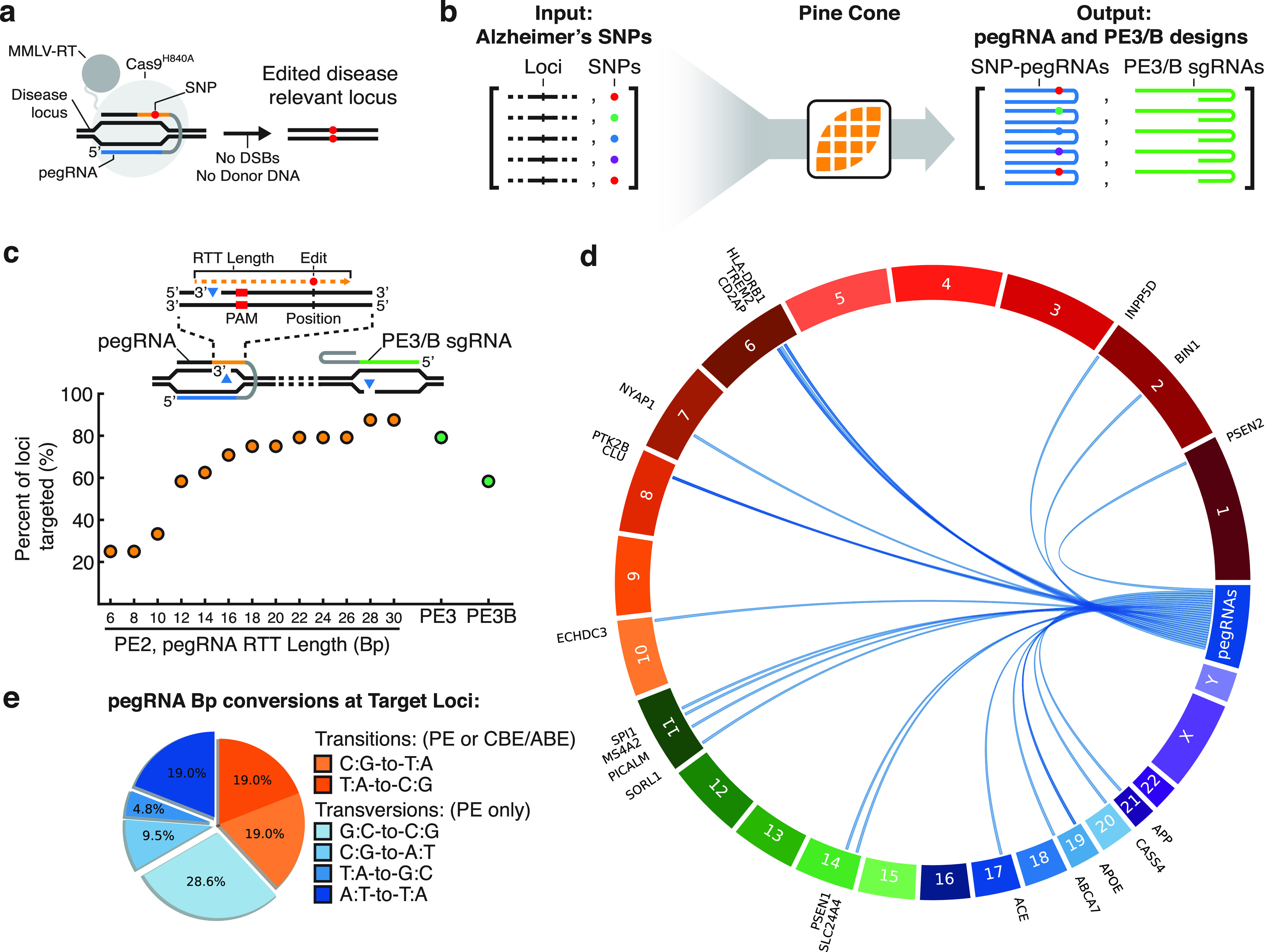
PINE-CONE design of pegRNAs of Alzheimer’s disease (AD)-related single nucleotide polymorphisms (SNPs). (a) Prime Editing mediated introduction of SNPs. A pegRNA targeting a disease locus encodes an edit, which is then incorporated into the target locus without the need for double-stranded DNA breaks (DSBs) or introduction of linearized donor DNA. (b) PINE-CONE rapidly analyzed and designed a library of pegRNAs and PE3 or PE3B sgRNAs for 24 AD-related loci. (c) The percent of loci targeted by pegRNAs was systematically analyzed for various RTT lengths. Inset schematic indicates valid edits that fall with the reverse transcription range. Longer RTT lengths expand the Prime Editing window and thus increase the number of targets up to 87% of loci with RTTs of 30 Bp (21/24). In parallel, PINE-CONE generated designs of PE3 sgRNAs for 79% of loci (19/24) and PE3B sgRNAs for 58% of loci (14/24). (d) PINE-CONE generated Circos mapping of pegRNAs to target loci indicates PINE-CONE successfully designs pegRNAs across numerous chromosome contexts. (e) A pie chart of PINE-CONE designed edits at the 24 AD-relevant loci. Transition mutations are accomplishable by cytosine base editors (CBEs), adenosine base editors (ABEs) and Prime Editors (PEs) (38%, orange). The majority of mutations consist of base transversion mutations (62%, blue).
