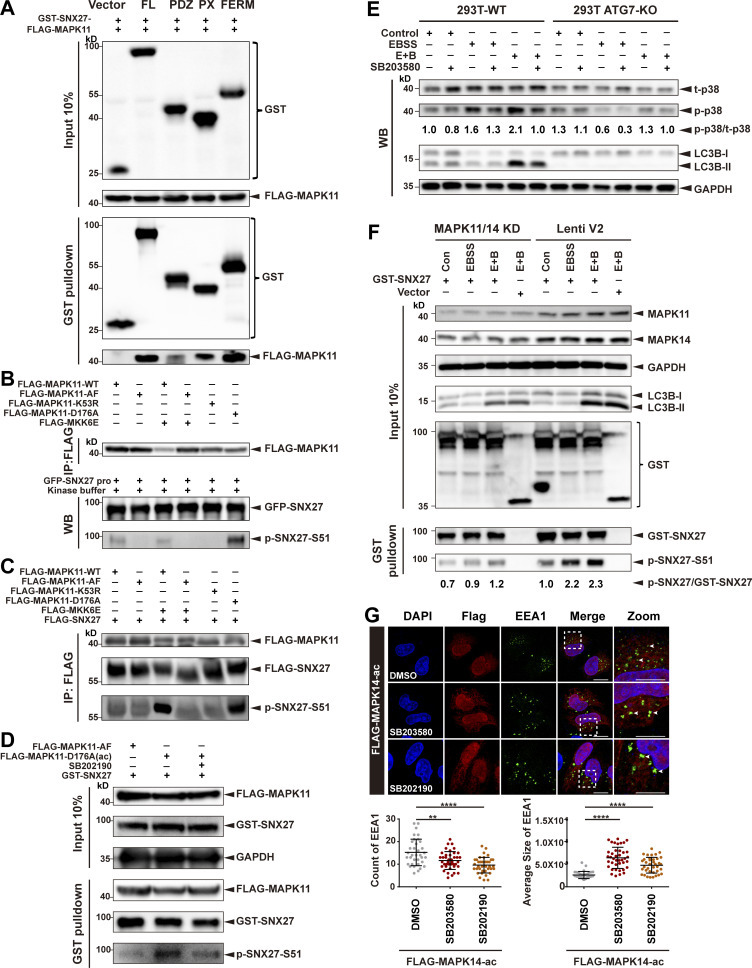
Figure S3.
MAPK11 phosphorylates SNX27 at Ser51 in vitro and in vivo. (A) Pull-down assay to assess interaction between MAPK11 and different domains of SNX27. HEK293T cells were cotransfected with FLAG-tagged MAPK11 and GST-tagged SNX27 FL, PDZ, PX, or FERM domain for 24 h. One tenth of the cell lysate was prepared as input, and the rest was used for pull-down with GSH affinity resin, followed by immunoblotting for GST-tagged SNX27 and FLAG-tagged MAPK11. Representative of three independent iterations. (B) MAPK11 phosphorylates SNX27 at Ser51 in vitro. HEK293T cells were transfected to FLAG-tagged MAPK11 WT, inactive MAPK11 (MAPK11-AF), inactive MAPK11 (MAPK11-K53R), or active MAPK11 (MAPK11-D176A), with or without FLAG-tagged active MKK6 (MKK6E), as indicated. MAPK11 was immunoprecipitated with anti-FLAG beads and mixed with purified GFP-tagged SNX27 and ATP. The assay was performed at 37°C and analyzed using the anti-phospho-SNX27-Ser51 antibody. Representative of three independent iterations. (C) MAPK11 phosphorylates SNX27 at Ser51 in vivo. HEK293T cells were cotransfected with FLAG-tagged SNX27 and various versions of FLAG-tagged MAPK14 as in B. Kinase and substrate proteins were immunoprecipitated with anti-FLAG beads, followed by immunoblotting using anti-FLAG and phospho-SNX27-Ser51 antibodies. Representative of three independent iterations. (D) In vivo MAPK11 kinase assay in the presence of kinase inhibitor (SB202190). HEK293 cells were cotransfected with GST-tagged SNX27 and FLAG-tagged MAPK11-AF, MAPK11-D176A, or MAPK11-D176A plus SB202190 (10 µM for 2 h). One tenth of the cell lysate was prepared as input, and the rest was used for pull-down with GSH resin, followed by immunoblotting for FLAG-MAPK11, GST-SNX27, and p-SNX27-Ser51. Representative of three independent iterations. (E) ATG7 deficiency decreases the phosphorylation of p38. WT and ATG7-KO HEK293T cells were starved in the presence or absence of p38 kinase inhibitor (SB203580, 10 µM for 2 h), followed by immunoblotting using antibodies against total p38, p-p38, LC3B, and GAPDH. Representative of two independent iterations. The numbers under the bands represent the p-p38/t-p38 ratio in each group. Band intensities were quantified with ImageJ and normalized to the 293T-WT control group (first lane). (F) MAPK11/14 depletion reduces phosphorylation of SNX27 at Ser51. HeLa cells were transfected with CRISPR-Cas9 plasmids targeting MAPK11 and MAPK14 (MAPK11/14 knockdown) or empty vector (Lenti V2). The knockdown and control cells were transfected with GST-tagged SNX27 for 24 h and starved for 4 h (E+B). One tenth of the cell lysate was prepared as input, and the rest was used for pull-down with GSH affinity resin, followed by immunoblotting for GST-SNX27 and p-SNX27-Ser51. Representative of two independent iterations. The numbers under the bands represent the p-SNX27/GST-SNX27 ratio in each group. Band intensities were quantified with ImageJ and normalized to the Lenti V2 control group. (G) Confocal imaging for EEA1 (green) puncta immunofluorescence staining of HeLa cells. Cells were transfected with FLAG-tagged MAPK14-D176A(ac) for 24 h and treated with 10 µM SB203580 or 10 µM SB 202190 for 2 h. Representative images from two independent experiments are shown. Scale bars: 10 µm (nonzoomed images) and 5 µm (zoomed images). Arrows indicate EEA1 puncta. The statistical analysis was performed based on the numbers and average sizes of the EEA1 puncta in each of the cells. Results for individual cells are plotted, along with the mean and SD for each group (n = 35 cells for DMSO, n = 40 for SB203580, and n = 38 for SB202190); P values were calculated using one-way ANOVA by Tukey’s HSD test or t test. ****, P < 0.0001; **, P < 0.01. HSD, honestly significant difference.