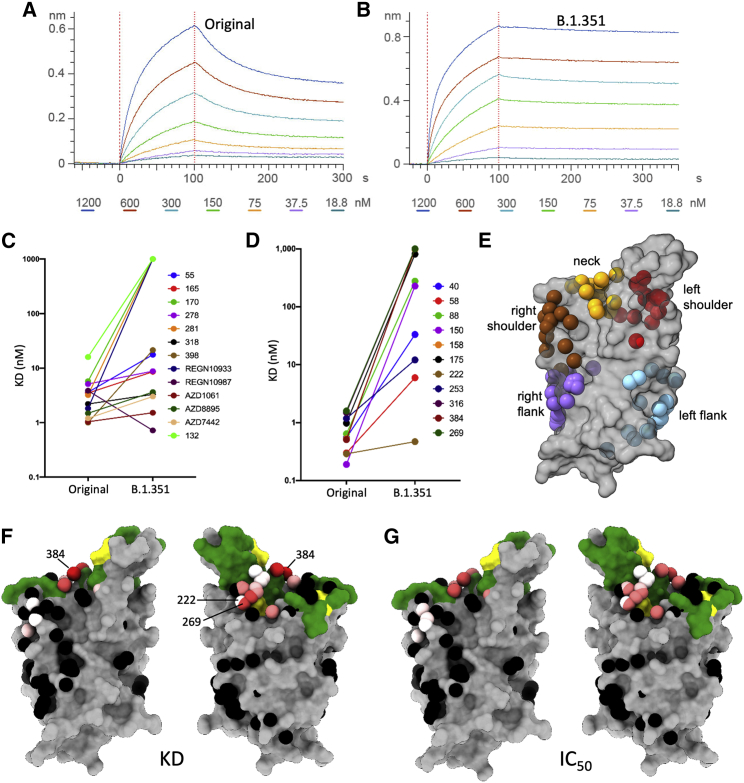
Figure 6.
Antibody RBD interaction and structural modeling
(A and B) BLI plots showing a titration series of binding to ACE2 (see STAR Methods) for (A) Wuhan RBD and (B) K417N, E484K, and N501Y B.1.351 RBD. Note the much slower off-rate for B.1.351.
(C and D) KD of RBD/mAb interaction measured by BLI for WT Wuhan RBD (left dots) and K417N, E484K, and N501Y B.1.351 RBD (right dots).
(E) Epitopes as defined by the clustering of mAbs on the RBD (gray).
(F) BLI data mapped onto the RBD using the method described in (Dejnirattisai et al., 2021). Front and back views of the RBD are depicted with the spheres representing the antibody binding sites colored according to the ratio (KDB.1.351/KDWuhan). For white, the ratio is 1; for red, it is <0.1 (i.e., at least 10-fold reduction). Black dots refer to mapped antibodies not included in this analysis; dark green to RBD ACE2-binding surface; and yellow to mutated K417N, E484K, and N501Y.
(G) As for the left pair, but colored according to the ratio of neutralization titers (half-maximal inhibitory concentration [IC50]B.1.351/[IC50]Victoria). For white, the ratio is 1; for red, it is <0.01 (i.e., at least 100-fold reduction). Note the strong concordance between the two effects, with 269 being the most strongly affected. The nearby pink antibodies are mainly the IGHV3-53 and IGHV3-66 antibodies.