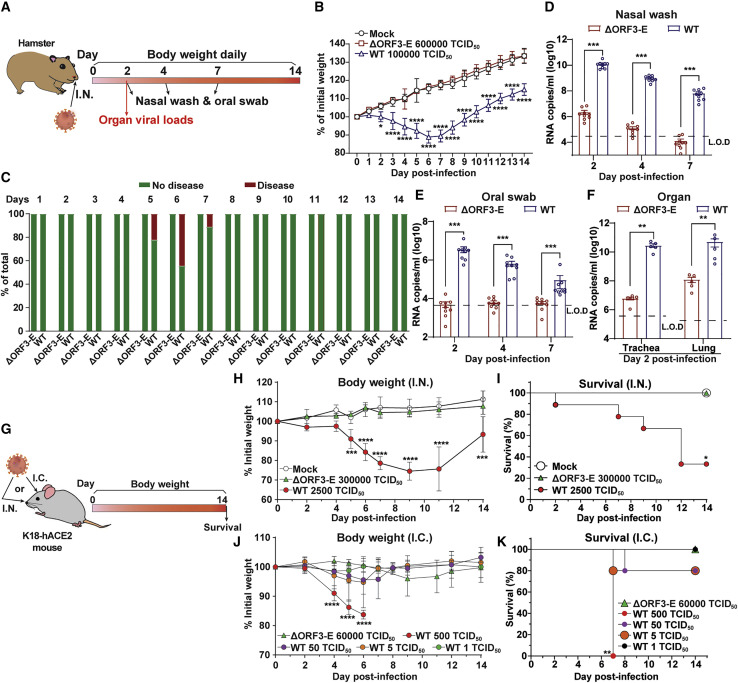
Figure 3.
Safety characterization of ΔORF3-E mNG virion in animal models
(A) Hamster experimental schedule. Four- to five-week-old male Syrian golden hamsters were intranasally (I.N.) inoculated with 105 TCID50 of WT SARS-CoV-2, 6 × 105 TCID50 of ΔORF3-E mNG virion, or PBS mock control. Hamsters were monitored for weight loss, disease, and viral RNA level.
(B) Hamster weight change (n = 9).
(C) Hamster disease (n = 9).
(D) Hamster nasal wash viral RNA level (n = 9).
(E) Hamster oral swab viral RNA level (n = 9).
(F) Viral RNA loads in hamster trachea and lung at day 2 post-infection (n = 5). Limit of detection (L.O.D.) was defined as the RNA copies detected from mock-infected hamster samples. The weight loss data are shown as mean ± standard deviation and statistically analyzed using two-way ANOVA Turkey’s multiple comparison. The genomic RNA levels are presented as mean ± standard error of the mean and analyzed by Mann-Whitney test. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
(G–K) Mouse experimental schedule. Seven- to nine-week-old K18-hACE2 mice were inoculated with WT SARS-CoV-2 or ΔORF3-E mNG virion via the I.N. or intracranial (I.C.) route. Mouse weight loss after I.N. (H) or I.C. (J) infection. Body weights were normalized to the initial weight. The means for each group (I.N.: WT SARS-CoV-2 [n = 9], ΔORF3-E mNG virion [n = 4], and mock [n = 4]; I.C.: WT SARS-CoV-2 500 TCID50 [n = 4], 50 TCID50 [n = 5], 5 TCID50 [n = 5], and 1 TCID50 [n = 5] and 6 × 104 TCID50 ΔORF3-E mNG virus [n = 4]) are indicated, with error bars indicating the standard deviation. A mixed-model ANOVA using Dunnett’s test for multiple comparisons was used to evaluate the statistical significance among groups: ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. Mouse survival after I.N. (I) and I.C. (K) inoculation was analyzed using the Gehan-Breslow-Wilcoxon test. Using groups with 100% survival as a comparator, a Bonferroni correction was applied manually to adjust the threshold for significance (indicated by ∗).