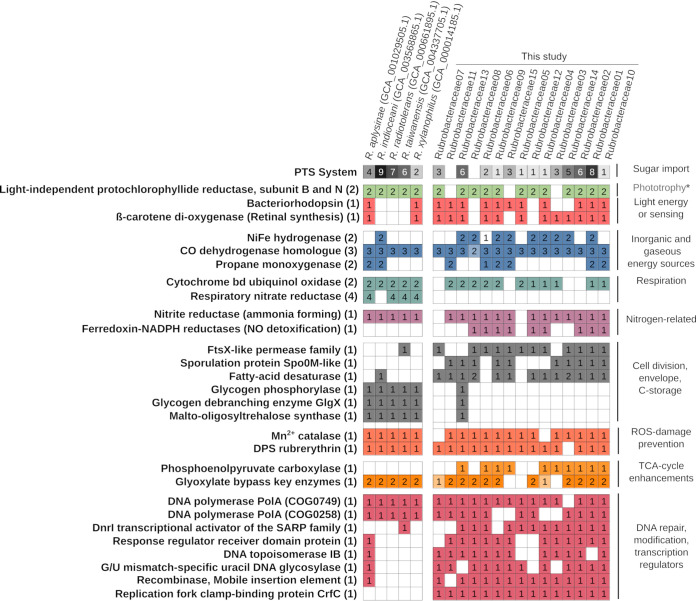
FIG 5.
Comparison of selected metabolic traits between Rubrobacteraceae MAGs from this study and sequenced genomes of Rubrobacteraceae isolates. The analysis is based on comparison of EggNOG orthologous groups detected in the genomes. In cases where the EggNOG/COG functional annotation was too general, annotations were completed based on specific RAST, Pfam, and UniProt search results for the given genes. The asterisk indicates that the genes encoding light-independent protochlorophyllide reductase subunits B and N do not represent a potential for phototrophy, as they are the only bacteriochlorophyll synthesis genes found in Rubrobacter genomes. The isolate genomes stem from aquatic environments, such as a Mediterranean sponge (Rubrobacter aplysineae) (57), Indian Ocean sediment (Rubrobacter indicoceanii) (58), mud and thermo-mineral water of a radioactive spring (Rubrobacter radiotolerans) (59), hot spring water, and thermally heated mud and soil (Rubrobacter taiwanensis) (60), and biofilm of a thermally polluted runoff of a carpet factory (Rubrobacter xylanophilus) (61).