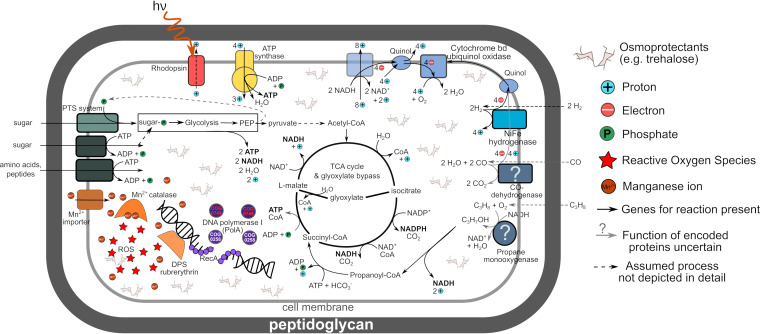
FIG 6.
Exemplary metabolic sketch of BSC Rubrobacteraceae species based on the most abundant MAG. The sketch illustrates basic energy generation and C acquisition mechanisms, as well as some stress resistance and survival mechanisms. Rubrobacteraceae encode genetic potential for a mixotrophic lifestyle with rhodopsin and a high-affinity hydrogenase to generate proton motive force. The exact functions of CO dehydrogenase and propane monooxygenase homologs are yet to be determined. The C metabolism is optimized for efficient C utilization by using phosphotransferase sugar import systems (PTS) and glyoxylate bypass of the TCA cycle. It is important to note the absence of terminal cytochrome c oxidase in the respiratory chain and exclusive reliance on the manganese-based catalase for combating reactive oxygen species (ROS). DNA-binding rubrerythrin might also contribute to preventing ROS damage to DNA. Also note the presence of an additional homolog of PolA polymerase (COG0749), key in double-stranded DNA break repair. For pathway evaluation, RAST annotations were analyzed with Pathway Tools v.24 (131), and missing enzymes, usually due to the absence of an EC number in the annotation or the use of an uncommon synonymous enzyme name, were searched manually in RAST and other annotations of the MAGs. The depicted pathways are simplified versions of pathway depictions in the MetaCyc database (132) used by Pathway Tools software.