ABSTRACT
Exposure to polychlorinated biphenyls (PCBs), an endocrine-disrupting compound, is ubiquitous despite decades-old bans on the manufacture and use of PCBs. Increased exposure to PCBs is associated with adverse health consequences throughout life, including type 2 diabetes and cancer. PCB exposure is also associated with alterations in epigenetic marks and gene transcription, which could lead to adverse health outcomes, but many of these are population-specific. To further investigate the association between PCB and epigenetic marks, DNA methylation was measured at 787,684 CpG sites in 641 peripheral blood samples from the Michigan Polybrominated Biphenyl (PBB) Registry. 1345 CpGs were associated with increased total PCB level after controlling for age, sex, and 24 surrogate variables (FDR < 0.05). These CpGs were enriched in active promoter and transcription associated regions (p < 0.05), and in regions around the binding sites for transcription factors involved in xenobiotic metabolism and immune function (FDR < 0.05). PCB exposure also associated with proportions of CD4T, NK, and granulocyte cell types, and with the neutrophil to lymphocyte ratio (NLR) (p < 0.05), and the estimated effect sizes of PCB on the epigenome were correlated with the effect sizes previously reported in an epigenome-wide study of C-reactive protein (r = 0.29; p = 2.22e-5), supporting previous studies on the association between PCB and immune dysfunction. These results indicate that PCB exposure is associated with differences in epigenetic marks in active regions of the genome, and future work should investigate whether these may mediate the association between PCB and health consequences.
KEYWORDS: Epigenetics, EWAS, epigenome-wide association study, EDC, endocrine-disrupting compound, immune function
Introduction
Exposure to polychlorinated biphenyls (PCBs) is ubiquitous. Despite decades-old bans on manufacture and use in the United States, biomonitoring studies have demonstrated that the general population has detectable levels of at least one of the 209 congeners of PCB [1,2]. Originally used as coolants and lubricants in electronic devices, PCBs are now recognized as an endocrine-disrupting compound (EDC), meaning that they can disrupt normal endocrine function by mimicking or blocking hormones [2]. Continued exposure to PCB happens primarily through dietary exposures such as eating contaminated fish, although it could also come from the small amounts of PCB in the soil and water or from contact with hazardous waste from PCB contamination sites [2]. Furthermore, PCBs tends to be biologically persistent with half-life estimates ranging from several years to over a decade [3–5], meaning that older people exposed to high levels of PCBs before the bans were introduced can still have high levels of PCBs circulating in their serum [2].
PCBs have been linked to a wide variety of health problems in both model systems and human cohorts. In rats and humans, PCB exposure is associated with thyroid disease and altered thyroid hormone levels, which can impact developing neurons [6–12]. PCB exposure is also associated with oestrogen-related health problems such as endometriosis, increased menstrual cycle length, later age of menarche, and increased risk for breast cancer [13–16]. Beyond associations with thyroid and oestrogen related hormones, PCB levels are also associated with an increased risk for common diseases such as type 2 diabetes [17], heart disease [18], liver disease [19], and cancer [20]. Additionally, increased exposure to PCBs is associated with immune-related phenotypes such as altered immune cell composition and increased inflammation [21,22]. Because of the wide variety of health problems associated with PCB exposure, their persistence in the environment, and the potential for ongoing exposure, PCBs remain a public health concern [23].
As part of the continued research into PCBs and human health, there has been increased research into the biological mechanisms by which PCBs can lead to adverse health outcomes. Studies on the effects of PCBs on reproductive organs have reported that PCBs can alter oestrogen receptor expression in ovarian follicles in chickens and affect sex steroid secretion by affecting the expression of steroidogenic genes [24]. Similarly, in rats, PCB exposure reduced testosterone, oestradiol, and androgen-binding protein levels in testicular interstitial fluid, potentially leading to the observed changes in testicular architecture [25]. PCB exposure has also been shown to increase the expression of inflammatory genes in the hypothalamus of rats and increase cytokines and C-reactive protein (CRP) – markers of inflammation – in endothelial cells by activating NF-κB signalling through altered epigenetic marks [26]. In human populations, PCBs have been associated with gene expression of immune-related genes in blood [27]. PCB exposure has also been associated with global DNA methylation levels in blood, but the direction has been inconsistent, potentially due to differences in exposure, population differences, and technologies [28–30]. In the only large epigenome-wide study of PCB exposure in humans published to-date, PCB exposure associated with 650 CpG sites with significant overlaps in the epigenetic signal from B-cell chronic lymphocyte leukaemia [31]. However, many of these were only significant in the men from one of the populations studied, suggesting that different populations may have different responses to PCB due to underlying genetic differences or differences in exposures, and that there may be sex-specific effects.
This study seeks to better understand the association between PCB exposure and DNA methylation levels in humans who were exposed to PCBs from typical environmental sources. To do this, we utilized the larger MethylationEPIC array and samples taken as part of the Michigan Polybrominated Biphenyl (PBB) Registry. Participants from this registry were recruited because they were believed to have high exposure to the structurally-related EDC, PBB, after an agricultural accident in the 1970s, and were used to study the association between PBB and DNA methylation [32,33]. However, PCB exposure has also been measured in this registry, and it is comparable to national exposure levels and has low correlation with their PBB exposure [1,34]. Therefore, this is a reasonable population to test for epigenetic associations with increasing exposure to PCB. We then utilized existing datasets and studies to annotate and interpret the epigenetic marks associated with PCB exposure. Finally, we attempted to replicate the previous CpGs associated with PCB [31] and tested whether there was any overlap between the CpGs previously reported to associate with PBB [32] and PCB. The results will improve our understanding of PCBs (and potentially other EDCs) and epigenetic marks, and help understand the health risks of people who are exposed.
Methods
Participant selection
Participants were selected from the Michigan PBB Registry. This cohort was started in 1976 by the Michigan Department of Community Health (MDCH; now the Michigan Department of Health and Human Services) after an agricultural accident contaminated livestock feed with PBB. The MDCH enrolled individuals believed to have had the highest exposure to PBB: families that lived on farms quarantined because of high PBB levels in livestock, people who ate food from quarantined farms, and chemical workers and their family members. Biological samples and health information from the original registry participants, their children, and other members of the community who were exposed to PBBs are still being collected by Emory University, which now manages the registry (http://pbbregistry.emory.edu/). Informed consent was obtained from each individual before participation and study protocols were approved by the Institutional Review Board at Emory University. Participants were previously selected from the registry for epigenetic analysis if they were exposed to PBB prior to being 50 years old, had given information on their health outcomes, had a recent (2004–2015) buffy coat or whole blood sample available for DNA extraction, and had detectable PBB levels in their serum (N = 666). All samples were taken between 2004–2015 (31–42 years after the exposure incident), but DNA methylation measurement occurred at one time to limit batch effects. PBB levels, PCB levels, lipid levels, and DNA methylation levels were all measured from fractions of the same blood sample collected from each participant. Current and past health and demographic information on all of these participants is still being collected.
Exposure assessment
While this population was originally recruited to study the effect of PBB exposure, PCB levels have also been measured in their serum. 209 possible congeners of PCB exist based on the number and position of the chlorine molecules around the biphenyl rings, but exposure to four common congeners of PCB (PCB-153, PCB-138, PCB-180, and PCB-118) was previously assessed in members of this registry using gas chromatography-tandem mass spectrometry [35]. The limit of detection (LOD) is 0.7 pg/mL for PCB-180; 1.6 pg/mL for PCB-153; 1.2 pg/mL for PCB-138; and 1.4 pg/mL for PCB-118. The extraction recovery ranged from 83.2 to 99.2%. The accuracy ranged from 89 to 119% and the precision ranged from 2.8 to 8.5%. Eighteen participants had PCB measurements that failed quality control checks and were excluded from all further analyses. For the purposes of this study, the value for congeners below the LOD in a sample was imputed as the LOD divided by the square root of 2 [36]. The congeners were summed to give a total PCB value per person. This was then transformed using a natural log so that the distribution was less skewed.
Lipid measurement
Total lipid level was calculated in this cohort as part of previous studies [6]. A Triglyceride Quantification Assay Kit (Abnova Corporation) was used to measure the total triglyceride content in serum, and a Cholesterol Assay Kit (Caymen Chemical Company) was used to measure total cholesterol content in serum. Both were done in accordance with the manufacturer’s recommendations. Total lipid amount was calculated based on these components as described elsewhere [37].
DNA extraction
As previously described [32], peripheral blood samples were collected from participants as part of the ongoing Michigan PBB Registry activities between 2004–2015. DNA was extracted from buffy coat samples using the QIAamp DNA Blood Mini Kit (QIAGEN, Hilden, Germany). DNA was extracted from the blood sample from which the exposure levels and lipid levels were assessed on to limit confounding.
MethylationEPIC Beadchip
As previously described [32], methylation levels were measured across the genome at 866,895 sites using the Infinium MethylationEPIC BeadChip (Illumina, San Diego, CA) [38]. Briefly, 1 microgram of DNA from participants’ buffy coats was bisulphite converted, amplified, fragmented, and hybridized to the BeadChip according to the manufacturer’s instructions. DNA from a female, lymphoblast line was used as a technical replicate. Data with low signal (detection p-value greater than 0.01) was considered missing, and samples and probes with missing data (missing in more than 10% of samples) were removed with CpGassoc [39]. Two samples were removed for low signal, and 5,982 probes were removed for missing data. Previously identified cross-reactive probes (N = 44,210) were also removed from the dataset [40]. BMIQ was used to adjust for probe type [41]. CpGs with gaps (5%) in their methylation proportions (indicative of a potential SNP) were identified using Gaphunter and removed [42]. 6 samples that were mismatches for sex, genetically identical to other samples, or flagged for sample contamination were removed. The 57 SNP probes were removed from the dataset. This resulted in a final dataset of 641 participants and 787,684 probes. For each individual sample at each probe, the methylation proportion (β) at that site was calculated from the methylated (M) and unmethylated (U) signal as β = M/(U+M). Significant surrogate variables were calculated using SVA to adjust for batch and cell type differences [43]. There were 24 significant surrogate variables, and they correlated with known confounders such as cell type composition (Figure S1). Cell type composition for multivariate analysis was calculated for each sample using Houseman’s method for estimating cell type proportion with methylation signals from CpGs that are distinct in blood cell types [44,45]. The DNA methylation data can be accessed on NCBI’s Gene Expression Omnibus (GSE116339).
Epigenome-wide association testing
Linear regression models in CpGassoc were used to interrogate the association between total PCB level and both the average methylation proportion across all probes that passed quality control (787,684 sites) and the methylation proportion at every probe individually [39]. Age at sample collection, sex, and the 24 significant surrogate variables estimated earlier were used as covariates. A Benjamini-Hochberg False Discovery Rate (FDR) of less than 0.05 was used to adjust for multiple testing [46]. As a sensitivity analysis, each congener of PCB was associated separately with DNA methylation, with age, sex, and 24 surrogate variables, and the results from this analysis were correlated with the results from the main analysis using a Pearson’s correlation coefficient. Additionally, a sensitivity analysis controlling for cell type composition and lipid levels (in addition to age, sex, and the 24 surrogate variables) was conducted and the results from this analysis were correlated with the results from the main analysis using a Pearson’s correlation coefficient. To identify potential sex-specific associations, the sample was also stratified by sex, and the association between PCB and DNA methylation was tested in each sex separately, adjusting for age and 24 surrogate variables. The association between global methylation (the average methylation proportion across all sites) and total PCB level was also tested using the same covariates. MissMethyl was used to determine whether any of the 330 KEGG pathways were enriched in the results to correct for the number of probes in each gene, and an FDR of less than 0.05 was used to adjust for multiple testing [47].
Enrichment tests for specific positional and functional regions
Chi-square tests were used to test for enrichment of PCB-associated CpGs in certain positions (based on location within a gene or proximity to a CpG island). CpG positions were determined from the annotation provided for each probe by the manufacturer. Functional regions were determined by the ChromHMM tracks publicly available on the UCSC Genome Browser [48–50]. Briefly, these tracks used data from multiple chromatin immunoprecipitation sequencing (ChIP-seq) experiments to annotate the genome into 25 categories based on the combination of histone marks seen in that region in that cell type. These 25 categories were then further condensed into 10 functional region types based on that category’s position relative to a gene, its function, and its effect on gene transcription. For the enrichment tests, the tracks from the most similar cell type to whole blood (the GM12878 lymphoblastoid cell line) were used. An alpha level of 0.05 was used to determine statistical significance for the positional and functional enrichment. Enrichment of transcription factor binding sites was tested using oPOSSUM-3, which is based on data from ChIP-seq data for various transcription factors. This reports a Z score for enrichment of transcription factor binding sites within 10 base pairs on either side of each CpG that was associated with PCB compared to 10 GC-matched CpGs that were not associated with PCB [51]. A Benjamini-Hochberg False Discovery Rate (FDR) of less than 0.05 was used to adjust for multiple testing.
Correlations with oestradiol and immune signalling
Because PCB is a known endocrine-disrupting compound and has been previously associated with oestrogen-related health conditions [15,16], we tested whether the epigenetic effect from oestradiol was correlated (using a Pearson’s correlation coefficient) with the epigenetic effect of PCB. This analysis included both the CpGs that were significantly associated with oestradiol (FDR < 0.05; N = 24,878) in a previous study [52] and the CpGs significantly associated with PCB in this study. Additionally, enrichment of oestradiol-associated CpGs in the CpGs significantly associated with PCB was tested using a chi-square test and results from the same previously published epigenome-wide study [52]. Because these were identified using data from the Infinium HumanMethylation450 BeadChip, the results from this study were first subset to the probes interrogated by both studies (N = 401,387).
Because PCB exposure has also been associated with inflammation and immune-related phenotypes [26,31], PCB exposure was associated with epigenetic-based immune phenotypes. First, the epigenetic signal from PCB was compared to the epigenetic profile of an immune phenotype estimated by a previously published epigenome-wide study of CRP (a marker of low-grade inflammation) [53]. Similar to the analysis with oestradiol, the effect sizes between the CpGs that overlap in both datasets and were significantly associated with CRP (FDR < 0.05; N = 206) were compared using a Pearson’s correlation coefficient [53]. Second, the estimated cell type proportions calculated earlier, as well as the neutrophil to lymphocyte ratio (calculated from the cell types estimated from DNA methylation) were regressed on total PCB, both in univariate analyses and controlling for age, sex, and lipid levels [54]. An alpha level of 0.05 was used to determine statistical significance.
Replication
We next sought to test 1) whether our results replicated the results of a previous epigenome-wide study with PCB [31], and 2) whether the epigenetic signal from PCB was similar to the epigenetic signal from the structurally similar PBB, as found in a previously published epigenome wide study [32]. To replicate the PCB-associated CpGs reported in the previous study, we tested whether there was correlation between the effect size reported with the CpGs significantly associated with PCB-156 in a Swedish male population (FDR < 0.01; the strongest association in that study [31]) and the effect size for total PCB in this study, both using the results from the combined-sex analysis and the males-only analysis. Additionally, for the congeners that were measured in both studies (PCB-180, PCB-138, PCB-153, and PCB-118), the effect size for the CpGs associated with each congener (FDR < 0.01) from the previous study and the effect size for that same congener from this study (combined-sex analysis and males-only analyses), were also tested for correlation. For testing the overlap between the epigenetic effect from PCB and the epigenetic effect of PBB, the effect sizes from both this analysis and the previously published analysis were compared with a correlation coefficient (including both the CpGs associated with PCB and the CpGs associated with PBB).
Results
Study population
The population of this study is composed of 641 people of primarily White/Non-Hispanic ancestry, which is representative of the population structure of rural Michigan in the 1970s. There were more female participants than male participants (N = 373 vs. 268). Participants had exposure to PBB due to an industrial accident (range: 0.01–236.73 ng/mL; Table 1) and exposure to PCB due to typical environmental exposure (range 0.07–8.11 ng/mL, Figure S2) [1,55]. There was low correlation between participants’ PCB and PBB exposure (r = 0.17; p = 5.43e-6), and a majority of participants (69.5%) had higher exposure to PCB than PBB. Current serum levels of all four measured PCB congeners were positively correlated (r = 0.65–0.99; Figure S3). Older age at sample collection was correlated with higher serum levels of PCB (r = 0.56; p < 2.2e-16), and males had significantly higher exposure than females (geometric mean of 0.86 vs. 0.67; p = 0.0007). Current PCB levels were not associated with current lipid level (r = 0.07; p = 0.06).
Table 1.
Descriptive characteristics of study population
| Demographics | |
|---|---|
| N | 641 |
| Percent malea | 268 (41.8%) |
| Current Age (years)b | 54.3 (12.8) |
| Total PCB level (ng/mL)c | 0.74 (2.55) |
| Total PCB level (ng/g lipid)c | 114.05 (2.61) |
| PCB-118 level (ng/mL)c | 0.04 (5.02) |
| PCB-118 level (ng/g lipid)c | 6.53 (4.92) |
| PCB-138 level (ng/mL)c | 0.22 (2.75) |
| PCB-138 level (ng/g lipid)c | 34.12 (2.81) |
| PCB-153 level (ng/mL)c | 0.25 (2.55) |
| PCB-153 level (ng/g lipid)c | 38.38 (2.61) |
| PCB-180 level (ng/mL)c | 0.19 (2.51) |
| PCB-180 level (ng/g lipid)c | 29.76 (2.58) |
| Total PBB level (ng/mL)c | 0.47 (4.65) |
| Total PBB level (ng/g lipid)c | 72.64 (4.90) |
| Race/Ethnicity a | |
| White/Non-Hispanic | 621 (97%) |
| White/Hispanic | 20 (3%) |
aFrequency and percentage
bMean and standard deviation
cGeometric mean and geometric standard error
PCB associates with DNA methylation
Similar to some studies of PCB exposure [28], global DNA methylation level was positively associated with total PCB, controlling for age, sex, and surrogate variables (t = 5.96, p = 4.14e-9). Out of the 787,684 CpG sites tested, current total PCB level associated with the methylation proportion at 1345 CpGs (FDR<.05; Figure 1, Table S1). Of these CpGs, 51.8% had higher methylation levels in those with higher levels of PCB (Figure S4). Results of the analysis that also adjusted for lipids and cell type proportions (in addition to age, sex, and the 24 surrogate variables) were highly correlated with results that just adjusted for age, sex, and 24 surrogate variables (R = 0.98; p < 2.2e-16; Figure S5). The effect sizes from the total PCB analysis were also highly correlated with the effect sizes from analyses of each congener individually (r = 0.63–0.98; Figure S6). When the analysis was conducted in males and females separately, 3 CpGs were associated with total PCB in males, including 2 CpGs not found significant in the combined sex analysis (Table S2), and 12 CpGs were associated with total PCB in females including 2 CpGs not found significant in the combined sex analysis (Table S3). The results from the 1349 CpGs associated with total PCB in any analysis were highly correlated in men and women (r = 0.83, p < 2.2e-16). However, the CpGs significant in the analysis only in females had a small effect in men and the CpGs significant in the analysis only in males had a small effect in females (Figure S7). Of the 1345 results significant in the main analysis, 1050 are annotated to 953 unique genes. The genes with the largest number of significant CpGs were GK (N = 7), TMEM187 (N = 6), and ITPKB (N = 5). Scatterplots for the most significant CpGs were plotted and showed no outliers driving the results (Figure 2). There was no enrichment of KEGG pathways among PCB-associated CpGs (Table S4). There was enrichment of PCB-associated CpGs in the 5ʹUTR, gene body, and exon band, but there was no enrichment in any island position (Figure S8).
Figure 1.
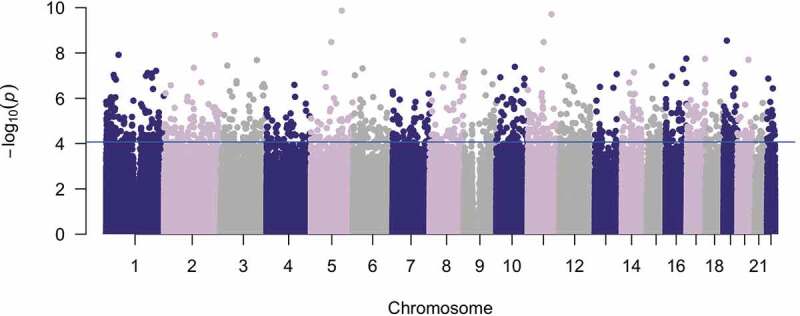
Current PCB levels associate with DNA methylation levels genome-wide. A Manhattan plot of the association of total PCB level with DNA methylation proportion at 787,684 sites. The x-axis is the location of each site across the genome. The y-axis is the – log10 of the p-value for the association with PCB. The blue line indicates statistical significance (FDR < 0.05). 1345 CpGs had a statistically significant association with total PBB level
Figure 2.
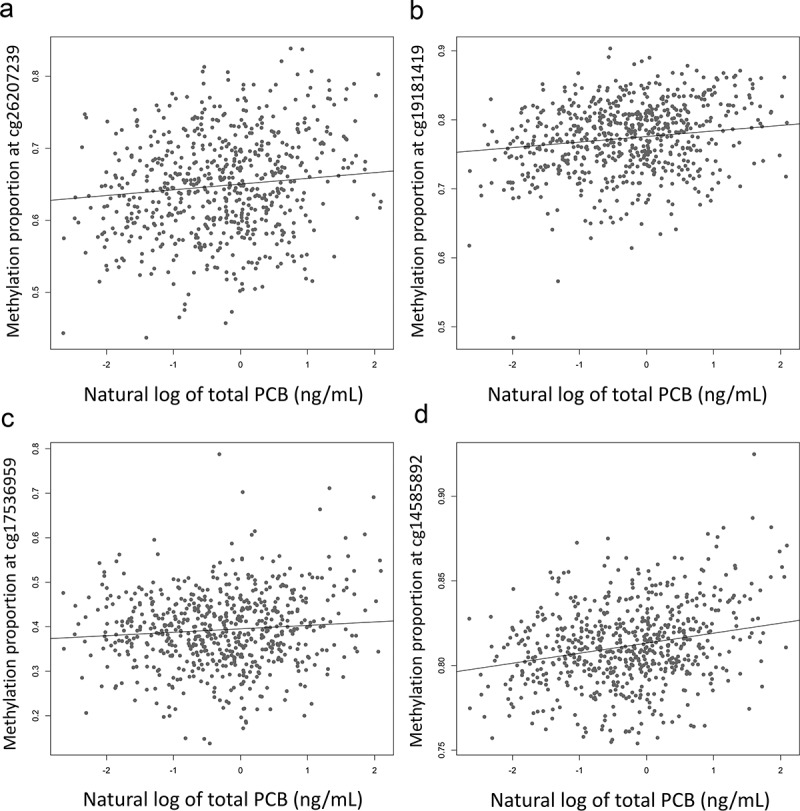
Scatterplots of the CpGs most highly associated with total PCB levels. The top CpGs most associated with total PCB: cg26207239 (BIRC2, p = 1.96e-10; (a), cg19181419 (p = 1.35e-10; (b), cg17536959 (p = 1.60e-9; (c), cg14585892 (LY6D, p = 2.81e-9; (d). The solid line is the regression of DNA methylation proportion at that CpG on total PCB level (controlling for age, sex and 24 surrogate variables)
Functional enrichment with PCB-associated CpGs
To better understand the function of the CpGs associated with PCB, they were annotated to functional regions as defined by the combination of chromatin marks. PCB-associated CpGs were depleted in regions that were inactive promoters (OR = 0.72, 95% CI = 0.52–0.98, p = 0.04), insulators (OR = 0.60, 95% CI = 0.41–0.85, p = 0.005), polycomb regions (OR = 0.60, 95% CI = 0.51–0.71, p = 2.43e-9), or heterochromatin regions (OR = 0.78, 95% CI = 0.67–0.91, p = 0.001), and they were enriched in regions that were active promoters (OR = 1.21, 95% CI = 1.06–1.38, p = 0.003) and associated with transcription (OR = 1.64, 95% CI = 1.43–1.87, p = 1.18e-13; Figure 3). Additionally, enrichment for transcription factor binding sites near the PCB-associated CpGs was also tested. Several transcription factor binding sites (N = 38) were enriched or depleted in the regions near PCB-associated CpGs (Table S5), with many of them being nuclear hormone receptors (HNF4A – p = 0.0003; PPARG::RXRA – p = 0.0006; NR2F1 – p = 0.009), involved in xenobiotic metabolism (HIF1A::ARNT – p = 7.09e-16; Arnt::Ahr – p = 1.39e-13; Arnt – p = 0.0003), or involved in immune response (NF-kappa B – p = 0.003).
Figure 3.
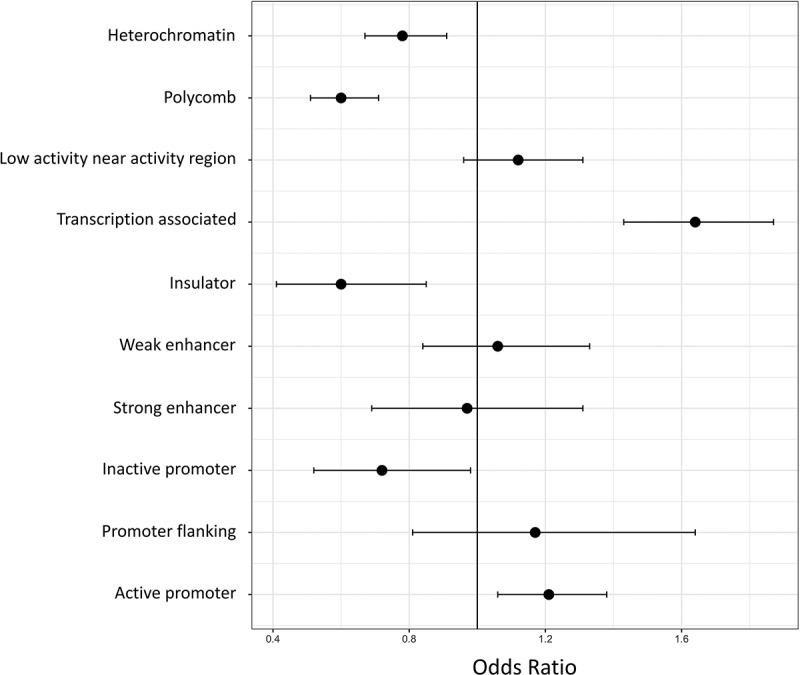
CpGs most associated with PCB are enriched in certain functional regions. Enrichment tests were conducted to determine if the CpGs most associated with PCB were enriched in certain functional regions. PCB-associated CpGs were enriched in transcription-associated regions and active promoters. They were depleted in polycomb, heterochromatin, insulator, and inactive promoter regions
Immune- and oestrogen-related associations with PCB-associated CpGs
Because PCB has been associated with endocrine disruption and immune-related cancers [13–16,26,31], it was of interest if the PCB-associated CpGs were among those previously associated with immune- or oestrogen-related phenotypes. To test for immune-related associations, we similarly investigated correlations between CpG-specific effect sizes from PCB and those previously estimated from CRP [53]. Among the set of CpGs associated with CRP (N =206), the effect from PCB and the effect from CRP were positively correlated (r = 0.29, p = 2.22e-5, Figure S9). We also found that, after adjustment for age, sex, and current lipid levels, PCB levels were positively associated with CD4 T and NK cell type proportions and negatively associated with the proportion of granulocytes and the neutrophil to lymphocyte ratio (NLR) (Table 2). When we tested for oestrogen-related associations, we found that the epigenetic profiles of CpG-specific effect sizes estimated for oestradiol [52] and for PCB were correlated, both when restricting analysis to oestradiol associated CpGs (N = 24,878, r = 0.14, p < 2.2e-16), and to PCB-associated CpGs (N = 664, r = 0.35, p < 2.2e-16; Figure S10) [52]. Of the 664 PCB-associated CpGs that were included in the oestradiol analysis, 103 of them were also significant in the oestradiol analysis (OR = 2.68; 95% CI = 2.16–3.29; p < 2.2e-16), suggesting that there is considerable overlap between the epigenetic marks that are associated with oestradiol levels and PCB.
Table 2.
Association of PCB with cell type proportions and NLR ratio, both in univariate analyses and controlled for age, sex, and lipid levels
| |
Univariate |
Multivariate |
||
|---|---|---|---|---|
| (95% CI) | P-value | (95% CI) | P-value | |
| CD8 T | −0.0057 (−0.0090, −0.0024) |
0.0007 | 0.0007 (−0.0031, 0.0046) |
0.70 |
| CD4 T | −0.0045 (−0.0097, 0.0005) |
0.08 | 0.0074 (0.0015, 0.0133) |
0.01 |
| B cells | −0.0008 (−0.0040, 0.0024) |
0.61 | 0.0008 (−0.0030, 0.0048) |
0.66 |
| NK | 0.0118 (0.0085, 0.0150) |
2.48e-12 | 0.0069 (0.0030, 0.0108) |
0.0005 |
| Monocytes | 0.0052 (0.0025, 0.0080) |
0.0001 | −0.0006 (−0.0037, 0.0025) |
0.69 |
| Granulocytes | 0.0001 (−0.0079, 0.0082) |
0.97 | −0.0099 (−0.0197, −0.0001) |
0.04 |
| log(NLR) | 0.0035 (−0.0445, 0.0516) |
0.88 | −0.0821 (−0.1394, −0.0248) |
0.004 |
Replication
We attempted both to replicate the epigenetic signal found in a previous epigenome-wide study of PCB [31] and to test the correlation between the epigenetic signal from PCB and the epigenetic signal from the structurally related PBB [32]. The strongest signal in the previous study was with PCB-156 (not measured in this population) in Swedish males. In the 536 CpGs that were significant (FDR < 0.01) in that study and passed quality control in this study, the effect sizes were not correlated in the combined-sex analysis (r = 0.04, p = 0.25), but had a low, significant correlation in the male-only analysis (r = 0.11, p = 0.006). For the four congeners (PCB-153, PCB-180, PCB-118, PCB-138) measured in both studies, the single-congener effect sizes for the 49 CpGs significant in the previous studies (FDR < 0.01) were negatively correlated with the single-congener effect sizes estimated for those CpGs in this study in the combined-sex analysis (r = −0.37, p = 0.007) and were not correlated with the single-congener effect sizes estimated for those CpGs in males only (r = −0.11, p = 0.41; Figure S11). The effect sizes for 1812 CpGs associated with PBB levels (FDR < 0.05 [32]) that also passed quality control in this study were weakly but significantly correlated with the effect size of those CpGs with PCB (r = 0.08, p = 0.0001; Figure S12(a)). The effect size for the 1345 CpGs associated with PCB levels was similarly correlated with the effect size of those CpGs with PBB (r = 0.08, p = 0.002; Figure S12(b)). While 5 CpGs were significantly associated with both PBB and PCB, the overlap was not significant (p = 0.27).
Discussion
This study investigated the association between PCB and DNA methylation through an epigenome-wide study of a population that was exposed to PCB at levels comparable to those considered background exposure in the United States. We then assessed the potential function of the associated CpGs by testing for enrichment of biological pathways, functional regions, and transcription factor binding sites, as well as testing for correlations between the epigenetic signal from PCB and the epigenetic signal from CRP and oestradiol. Finally, we attempted to replicate previously published CpGs associated with PCB and test whether the CpGs that associate with the structurally-similar PBB overlap with the CpGs that associate with PCB. In this study, 1345 CpGs associated with PCBs, after adjusting for age, sex, and 24 surrogate variables. These results were consistent if estimated cell type proportions were also controlled for, indicating that these results are robust to different methods of adjustment for heterogeneity, and were consistent with results from analyses of each congener individually, indicating that the method for summing the congeners did not bias the results. Slightly over half of these CpGs (51.8%) had a higher proportion of DNA methylation with higher current levels of PCB. We also found that higher PCB was associated with higher global methylation levels. This is consistent with some [28], but not all [29,30] previous studies, although some of this inconsistency may certainly be due to differences in technologies. Finding an association between PCB and specific sites in the epigenome is also consistent with a previous study and there was a positive association between the effect sizes from their reported results and the results from analysis of total PCB in just the males from this study [31], but the same sites were not associated in these two studies, and the effect size for the CpGs reported in that study were not positively correlated with the effect size for those CpGs in the combined-sex analysis from this study. The previously reported CpGs were only significantly associated with PCB in the Swedish men and were not associated in the Italian men or the women who were studied. Since this study involved presumably mixed European ancestry, there could be population-specific differences in response to PCBs, either due to underlying genetic differences or differences in PCB congeners and exposure routes, that explain the inconsistent findings in these two studies. Additionally, there might be a sex-specific effect of PCB on the epigenome based on previously reported studies [31] and the results from this study. While the correlation of CpGs associated with total PCB was high between men and women, there were 12 CpGs associated in the female-only analysis that had a small effect in the male-only analysis, as well as 3 CpGs associated in the male-only analysis that had a small effect in the female-only analysis. However, given the large number of covariates that had to be adjusted for in this analysis, it is likely that larger sample sizes are needed to further explore the sex-specific effects of PCB exposure.
While no biological pathways were enriched, these results still could have a functional role. We found that the PCB-associated CpGs were more likely to be in active promoter and transcription associated regions of the epigenome. Additionally, they were enriched in the regions near the binding sites for nuclear hormone receptors and transcription factors involved in xenobiotic metabolism and immune function. Many of these functional enrichments, for example finding NF-κB and AHR binding sites enriched in the results, are what is expected from exposure to a dioxin-like EDC and are consistent with previous reports in in vitro models [26,56], and taken together, suggest that these molecules are bound and activated in response to PCBs. These results also suggest that PCB could be impacting gene expression, and therefore cellular function, since the associated CpGs are near transcribed regions of the genome, consistent with previous studies [27]. However, future studies would be needed to directly test whether the associated CpGs are associated with any cellular phenotypes or health consequences.
The results from this study also support PCB having an effect on the immune system. We found that the effect size for CpGs associated with the inflammatory marker CRP was correlated with the effect size from PCB. This supports our other result that NF-κB binding sites were enriched near our PCB-associated CpGs, since NF-κB is involved in inflammatory response. Additionally, we found that PCB was associated with certain cell type proportions and NLR, even after adjusting for age, sex, and lipid levels. However, it associated with lower, not higher, NLR, when a higher NLR is indicative of inflammation. Therefore, the epigenetic effects of PCB exposure seem to have some correlations with inflammation and immune function, similar to other reported studies [14,26,31]. It is also consistent with previous studies that have found increased exposure to PCBs is associated with decreased thymic volume, altered antibody production, and an increased number of lymphocytes and T-cells [57–59]. However, not all of the studies have had consistent, clinically relevant health outcomes, and more studies are needed to test whether typical exposure to PCB causes alterations to immune function and whether alterations to epigenetic marks mediate any potential immune-related health outcomes.
In this study, the effect that PCB had on the epigenome was positively correlated with that reported for oestradiol, a natural oestrogen [52]. Additionally, a significant number of the CpGs associated with PCB are also associated with oestradiol levels. Since PCB has previously been shown to affect oestradiol levels by altering the expression of steroidogenic genes and to associate with oestrogen-related health outcomes like age of menarche, cycle length, and breast cancer risk [13,15,16,24], this supports that PCB may be impacting oestrogen function and thus have wide impact on health. Because this is a cross-sectional study, we cannot distinguish between PCB affecting oestrogen levels, which then affects oestrogen-associated CpGs, or if PCB acts similarly to an oestrogen on the same CpGs, or some combination of the two. However, either would indicate that PCB affects oestrogen signalling. Future studies into the mechanism by which PCB affects oestrogen signalling and potential oestrogen-related health outcomes are warranted.
We also found that, despite the structural similarity between PCB and PBB, there was very low correlation between their epigenetic signal, and only 5 CpGs were significant in both analyses. First, this suggests that our PCB results are not driven by the high PBB exposure in some participants. Additionally, this suggests that these molecules may have different effects on cellular function, and therefore, different health risks despite their structural similarity. While both PCB and PBB do increase risk for certain adverse health outcomes, such as breast cancer [16,60], they also have opposite effects on other outcomes, such as age of menarche [15,61]. It is possible that the CpGs that differ between the two EDCs could mediate these health outcomes that are different between the two molecules. However, given the large effect that PBB had on the epigenome [32], it is possible that some of the surrogate variables may have accounted for the larger PBB exposure in some individuals and thus absorbed the variation in CpGs that would be shared by PBB and PCB. This would have limited our ability to detect these shared CpGs in this study. However, using the surrogate variables in this analysis was necessary to adjust for potential unmeasured confounders like smoking status and BMI.
Despite the limited overlap between CpGs associated with PBB and PCB, it is important to note that the correlation between PBB’s and PCB’s epigenetic signal was still statistically significant. Additionally, some of the transcription factors associated with PBB, for example, HIF1A::ARNT, were significant in this study, and transcription factors involved in xenobiotic metabolism and nuclear hormone receptors (even if they were different transcription factors) were significant in both studies [32]. The effect sizes from both molecules were also positively associated with the effect sizes from CRP and oestradiol. Therefore, both PBB and PCB could still be associated with the same biological pathways (and thus some of the same health outcomes) even if they are associated with different CpGs. Some of the similarities and the differences between PCB and PBB may also be due to combinations of exposures that have different effects than either chemical individually. Since any organism has multiple environmental exposures, developing methods that can analyse combinations of exposures on an epigenome-wide scale is needed. Overall, more research is needed to determine the mechanisms that mediate the response to both PBB and PCB exposure to better understand the effect each has on cellular function and health.
The results of this study should be interpreted in light of its limitations. While the PCB exposure levels were comparable to background exposure levels for the general population in the United States, this study population was predominantly composed of people who self-report as white/non-Hispanic. Given that genetic differences could influence response to PCB, it is not known if these 1345 CpGs would associate with PCB exposure in a population with a different genetic ancestry. Therefore, more research is needed on the epigenetic effects of PCB in more diverse cohorts. Additionally, DNA methylation was only assessed in blood samples, and so it is not known if PCB and epigenetic marks would associate in other relevant tissues. Also, not enough participants reported health information on potential confounders and conditions, such as BMI. This meant that we instead adjusted for 24 surrogate variables, which may have decreased our power to detect associations at some CpGs, particularly in the stratified analyses. Furthermore, this did not allow us to investigate the potential for these CpGs to mediate risk for certain health conditions. For this to be done, studies with larger sample sizes with more detailed phenotyping are needed.
However, this study did have several strengths. By incorporating all significant surrogate variables into the linear regression models, we were able to adjust for both known confounders (such as differences in cell type proportions or batch effects) and unknown confounders (such as smoking or BMI, which was not available for all participants). This is also the largest epigenome-wide study of PCB to-date, and our use of the newer MethylationEPIC BeadChip enabled wider coverage of the epigenome and a greater focus on enhancer regions than the older arrays or global methylation methods. We were also able to measure PCBs and DNA methylation levels in the same tissue to also limit potential confounding, Finally, we were able to use existing datasets and studies to better understand the potential function of the CpGs associated with PCB.
In conclusion, PCBs were associated with the proportion of DNA methylation at 1345 CpGs. While they were not enriched in any particular biological pathways, these CpGs were more likely to be in active promoters and transcribed regions, suggesting they may have a functional effect. Additionally, they were enriched in the binding sites for transcription factors that were nuclear hormone receptors or involved in xenobiotic metabolism and immune function. The epigenetic signal from PCB was also associated with the epigenetic signal from oestradiol and CRP. Taken together, these results suggest typical levels of exposure to PCB affect specific sites in the epigenome consistent with an EDC and may affect immune function, consistent with previous studies of PCB.
Supplementary Material
Acknowledgments
We are grateful to the members of the Michigan PBB Registry for their participation and engagement with research studies over the past 40 years, to the Michigan Department of Health and Human Services, which had the foresight to create the Registry, and to our community partners (PBB Citizen Advisory Board, Pine River Superfund Citizen Task Force, and the Mid-Michigan District Health Department) who continue to provide guidance and insight to the Michigan PBB Research.
Funding Statement
This work was supported by the National Institute of Environmental Health Sciences [NIEHS; R01ES024790, R01ES025775, R24ES028528, P30ES019776] and the National Institute of General Medical Sciences [T32GM008490]. This study was supported in part by the Emory Integrated Genomics Core (EIGC), which is subsidized by the Emory University School of Medicine and is one of the Emory Integrated Core Facilities. Additional support was provided by the Georgia Clinical & Translational Science Alliance of the National Institutes of Health under Award Number [UL1TR002378]. The content is solely the responsibility of the authors and does not necessarily reflect the official views of the National Institutes of Health.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Xue J, Liu SV, Zartarian VG, et al. Analysis of NHANES measured blood PCBs in the general US population and application of SHEDS model to identify key exposure factors. J Expo Sci Environ Epidemiol. 2014;24(6):615–621. PubMed PMID: 24424407 [DOI] [PubMed] [Google Scholar]
- [2].ATSDR . Polychlorinated Biphenyls (PCBs) toxicity: what are adverse health effects of PCB exposure? Atlanta GA, editor. Atlanta (GA): US Department of Health and Human Services PHS; 2018. http://www.atsdr.cdc.gov/csem/pcb/docs/pcb.pdf. [Google Scholar]
- [3].Grandjean P, Budtz-Jorgensen E, Barr DB, et al. Elimination half-lives of polychlorinated biphenyl congeners in children. Environ Sci Technol. 2008;42(18):6991–6996. Epub 2008/10/16.PubMed PMID: 18853821; PMCID: PMC2600453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Karmaus W, Fussman C, Muttineni J, et al. Backward estimation of exposure to organochlorines using repeated measurements. Environ Health Perspect. 2004;112(6):710–716. Epub 2004/05/04. PubMed PMID: 15121515; PMCID: PMC1241966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ritter R, Scheringer M, MacLeod M, et al. Intrinsic human elimination half-lives of polychlorinated biphenyls derived from the temporal evolution of cross-sectional biomonitoring data from the United Kingdom. Environ Health Perspect. 2011;119(2):225–231. Epub 2010/ 10/12. PubMed PMID: 20934951; PMCID: PMC3040610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Jacobson MH, Darrow LA, Barr DB, et al. Serum polybrominated biphenyls (PBBs) and polychlorinated biphenyls (PCBs) and thyroid function among Michigan adults several decades after the 1973–1974 PBB contamination of livestock feed. Environ Health Perspect. 2017;125(9):097020. Epub 2017/09/28. PubMed PMID: 28953452; PMCID: PMC Journal - In Process [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Miller VM, Sanchez-Morrissey S, Brosch KO, et al. Developmental coexposure to polychlorinated biphenyls and polybrominated diphenyl ethers has additive effects on circulating thyroxine levels in rats. Toxicol Sci. 2012;127(1):76–83. PubMed PMID: 22345314; PMCID: PMC3327870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Byrne JJ, Carbone JP, Hanson EA.. Hypothyroidism and abnormalities in the kinetics of thyroid hormone metabolism in rats treated chronically with polychlorinated biphenyl and polybrominated biphenyl. Endocrinology. 1987;121(2):520–527. PubMed PMID: 3036477 [DOI] [PubMed] [Google Scholar]
- [9].Gauger KJ, Giera S, Sharlin DS, et al. Polychlorinated biphenyls 105 and 118 form thyroid hormone receptor agonists after cytochrome P4501A1 activation in rat pituitary GH3 cells. Environ Health Perspect. 2007;115(11):1623–1630. PubMed PMID: 18007995; PMCID: PMC2072832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].You SH, Gauger KJ, Bansal R, et al. 4-Hydroxy-PCB106 acts as a direct thyroid hormone receptor agonist in rat GH3 cells. Mol Cell Endocrinol. 2006;257–258:26–34. PubMed PMID: 16930818 [DOI] [PubMed] [Google Scholar]
- [11].Turyk ME, Anderson HA, Persky VW.. Relationships of thyroid hormones with polychlorinated biphenyls, dioxins, furans, and DDE in adults. Environ Health Perspect. 2007;115(8):1197–1203. PubMed PMID: 17687447; PMCID: PMC1940071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Bansal R, You SH, Herzig CT, et al. Maternal thyroid hormone increases HES expression in the fetal rat brain: an effect mimicked by exposure to a mixture of polychlorinated biphenyls (PCBs). Brain Res Dev Brain Res. 2005;156(1):13–22. PubMed PMID: 15862623 [DOI] [PubMed] [Google Scholar]
- [13].Buck Louis GM, Rios LI, McLain A, et al. Persistent organochlorine pollutants and menstrual cycle characteristics. Chemosphere. 2011;85(11):1742–1748. Epub 2011/10/25. PubMed PMID: 22018858; PMCID: PMC4143889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Huang Q, Chen Y, Chen Q, et al. Dioxin-like rather than non-dioxin-like PCBs promote the development of endometriosis through stimulation of endocrine-inflammation interactions. Arch Toxicol. 2017;91(4):1915–1924. Epub 2016/ 09/25. PubMed PMID: 27663891 [DOI] [PubMed] [Google Scholar]
- [15].Attfield KR, Pinney SM, Sjodin A, et al. Longitudinal study of age of menarche in association with childhood concentrations of persistent organic pollutants. Environ Res. 2019;176:108551. Epub 2019/08/04. PubMed PMID: 31376650 [DOI] [PubMed] [Google Scholar]
- [16].Morgan M, Deoraj A, Felty Q, et al. Environmental estrogen-like endocrine disrupting chemicals and breast cancer. Mol Cell Endocrinol. 2017;457:89–102. Epub 2016/ 10/19. PubMed PMID: 27717745 [DOI] [PubMed] [Google Scholar]
- [17].Wolf K, Bongaerts BWC, Schneider A, et al. Persistent organic pollutants and the incidence of type 2 diabetes in the CARLA and KORA cohort studies. Environ Int. 2019;129:221–228. Epub 2019/05/28. PubMed PMID: 31132656 [DOI] [PubMed] [Google Scholar]
- [18].Akesson A, Donat-Vargas C, Berglund M, et al. Dietary exposure to polychlorinated biphenyls and risk of heart failure - A population-based prospective cohort study. Environ Int. 2019;126:1–6. Epub 2019/02/19. PubMed PMID: 30776745 [DOI] [PubMed] [Google Scholar]
- [19].Clair HB, Pinkston CM, Rai SN, et al. Liver disease in a residential cohort with elevated polychlorinated biphenyl exposures. Toxicol Sci. 2018;164(1):39–49. Epub 2018/04/24. PubMed PMID: 29684222; PMCID: PMC6016643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Cao J, Fan T, Li W, et al. Association study between plasma levels of polychlorinated biphenyls and risk of cutaneous malignant melanoma. Environ Int. 2019;126:298–301. Epub 2019/03/03. PubMed PMID: 30825748 [DOI] [PubMed] [Google Scholar]
- [21].Haase H, Fahlenkamp A, Schettgen T, et al. Immunotoxicity monitoring in a population exposed to polychlorinated biphenyls. Int J Environ Res Public Health. 2016;13(3):295. Epub 2016/03/24. PubMed PMID: 27005643; PMCID: PMC4808958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Petriello MC, Brandon JA, Hoffman J, et al. Dioxin-like PCB 126 increases systemic inflammation and accelerates atherosclerosis in lean LDL receptor-deficient mice. Toxicol Sci. 2018;162(2):548–558. Epub 2017/12/08. PubMed PMID: 29216392; PMCID: PMC5888982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gore AC, Chappell VA, Fenton SE, et al. EDC-2: the endocrine society’s second scientific statement on endocrine-disrupting chemicals. Endocr Rev. 2015;36(6):E1–E150. PubMed PMID: 26544531; PMCID: PMC4702494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sechman A, Batoryna M, Antos PA, et al. Effects of PCB 126 and PCB 153 on secretion of steroid hormones and mRNA expression of steroidogenic genes (STAR, HSD3B, CYP19A1) and estrogen receptors (ERalpha, ERbeta) in prehierarchical chicken ovarian follicles. Toxicol Lett. 2016;264:29–37. Epub 2016/ 11/12. PubMed PMID: 27832956 [DOI] [PubMed] [Google Scholar]
- [25].Sugantha Priya E, Sathish Kumar T, Balaji S, et al. Lactational exposure effect of polychlorinated biphenyl on rat Sertoli cell markers and functional regulators in prepuberal and puberal F1 offspring. J Endocrinol Invest. 2017;40(1):91–100. Epub 2016/09/12. PubMed PMID: 27614457 [DOI] [PubMed] [Google Scholar]
- [26].Liu D, Perkins JT, Petriello MC, et al. Exposure to coplanar PCBs induces endothelial cell inflammation through epigenetic regulation of NF-kappaB subunit p65. Toxicol Appl Pharmacol. 2015;289(3):457–465. Epub 2015/11/01. PubMed PMID: 26519613; PMCID: PMC4662647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Leijs MM, Gan L, De Boever P, et al. Altered gene expression in dioxin-like and non-dioxin-like PCB exposed peripheral blood mononuclear cells. Int J Environ Res Public Health. 2019;16(12):2090. Epub 2019/06/16. PubMed PMID: 31200452; PMCID: PMC6617415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Lind L, Penell J, Luttropp K, et al. Global DNA hypermethylation is associated with high serum levels of persistent organic pollutants in an elderly population. Environ Int. 2013;59:456–461. PubMed PMID: 23933504 [DOI] [PubMed] [Google Scholar]
- [29].Rusiecki JA, Baccarelli A, Bollati V, et al. Global DNA hypomethylation is associated with high serum-persistent organic pollutants in Greenlandic Inuit. Environ Health Perspect. 2008;116(11):1547–1552. PubMed PMID: 19057709; PMCID: PMC2592276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kim KY, Kim DS, Lee SK, et al. Association of low-dose exposure to persistent organic pollutants with global DNA hypomethylation in healthy Koreans. Environ Health Perspect. 2010;118(3):370–374. PubMed PMID: 20064773; PMCID: PMC2854765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Georgiadis P, Gavriil M, Rantakokko P, et al. DNA methylation profiling implicates exposure to PCBs in the pathogenesis of B-cell chronic lymphocytic leukemia. Environ Int. 2019;126:24–36. Epub 2019/02/19. PubMed PMID: 30776747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Curtis SW, Cobb DO, Kilaru V, et al. Exposure to polybrominated biphenyl (PBB) associates with genome-wide DNA methylation differences in peripheral blood. Epigenetics. 2019;1–15. DOI: 10.1080/15592294.2019.1565590. PubMed PMID: 30676242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Curtis SW, Cobb DO, Kilaru V, et al. Exposure to polybrominated biphenyl and stochastic epigenetic mutations: application of a novel epigenetic approach to environmental exposure in the Michigan polybrominated biphenyl registry. Epigenetics. 2019;14(10):1003–1018. Epub 2019/ 06/16. PubMed PMID: 31200609; PMCID: PMC6691996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Chang CJ, Terrell ML, Marcus M, et al. Serum concentrations of polybrominated biphenyls (PBBs), polychlorinated biphenyls (PCBs) and polybrominated diphenyl ethers (PBDEs) in the Michigan PBB registry 40 years after the PBB contamination incident. Environ Int. 2020;137:105526. Epub 2020/ 02/18. PubMed PMID: 32062441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Marder ME, Panuwet P, Hunter RE, et al. Quantification of polybrominated and polychlorinated biphenyls in human matrices by isotope-dilution gas chromatography-tandem mass spectrometry. J Anal Toxicol. 2016;40(7):511–518. PubMed PMID: 27445313; PMCID: PMC4986627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Helsel DR. Less than obvious - statistical treatment of data below the detection limit. Environ Sci Technol. 1990;24(12):1766–1774. [Google Scholar]
- [37].Phillips DL, Pirkle JL, Burse VW, et al. Chlorinated hydrocarbon levels in human serum: effects of fasting and feeding. Arch Environ Contam Toxicol. 1989;18(4):495–500. PubMed PMID: 2505694 [DOI] [PubMed] [Google Scholar]
- [38].Pidsley R, Zotenko E, Peters TJ, et al. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol. 2016;17(1):208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Barfield RT, Kilaru V, Smith AK, et al. CpGassoc: an R function for analysis of DNA methylation microarray data. Bioinformatics. 2012;28(9):1280–1281. PubMed PMID: 22451269; PMCID: PMC3577110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].McCartney DL, Walker RM, Morris SW, et al. Identification of polymorphic and off-target probe binding sites on the Illumina infinium MethylationEPIC BeadChip. Genom Data. 2016;9:22–24. PubMed PMID: 27330998; PMCID: PMC4909830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Teschendorff AE, Marabita F, Lechner M, et al. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013;29(2):189–196. PubMed PMID: 23175756; PMCID: PMC3546795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Andrews SV, Ladd-Acosta C, Feinberg AP, et al. “Gap hunting” to characterize clustered probe signals in Illumina methylation array data. Epigenetics Chromatin. 2016;9:56. Epub 2016/12/17. PubMed PMID: 27980682; PMCID: PMC5142147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Leek JT, Storey JD. A general framework for multiple testing dependence. Proc Natl Acad Sci U S A. 2008;105(48):18718–18723. Epub 2008/11/27. PubMed PMID: 19033188; PMCID: PMC2586646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. PubMed PMID: 22568884; PMCID: PMC3532182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10):R115. PubMed PMID: 24138928; PMCID: PMC4015143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc. 1995;57(1):289–300. [Google Scholar]
- [47].Phipson B, Maksimovic J, Oshlack A. missMethyl: an R package for analyzing data from Illumina’s humanmethylation450 platform. Bioinformatics. 2016;32(2):286–288. PubMed PMID: 26424855 [DOI] [PubMed] [Google Scholar]
- [48].Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods. 2012;9(3):215–216. PubMed PMID: 22373907; PMCID: PMC3577932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol. 2010;28(8):817–825. PubMed PMID: 20657582; PMCID: PMC2919626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Hoffman MM, Ernst J, Wilder SP, et al. Integrative annotation of chromatin elements from ENCODE data. Nucleic Acids Res. 2013;41(2):827–841. PubMed PMID: 23221638; PMCID: PMC3553955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kwon AT, Arenillas DJ, Worsley Hunt R, et al. oPOSSUM-3: advanced analysis of regulatory motif over-representation across genes or ChIP-Seq datasets. G3 (Bethesda). 2012;2(9):987–1002. PubMed PMID: 22973536; PMCID: PMC3429929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Maddox SA, Kilaru V, Shin J, et al. Estrogen-dependent association of HDAC4 with fear in female mice and women with PTSD. Mol Psychiatry. 2018;23(3):658–665. PubMed PMID: 28093566; PMCID: PMC5513798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Ligthart S, Marzi C, Aslibekyan S, et al. DNA methylation signatures of chronic low-grade inflammation are associated with complex diseases. Genome Biol. 2016;17(1):255. Epub 2016/ 12/14. PubMed PMID: 27955697; PMCID: PMC5151130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Koestler DC, Usset J, Christensen BC, et al. DNA methylation-derived neutrophil-to-lymphocyte ratio: an epigenetic tool to explore cancer inflammation and outcomes. Cancer Epidemiol Biomarkers Prev. 2017;26(3):328–338. PubMed PMID: 27965295; PMCID: PMC5336518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Sjodin A, Wong LY, Jones RS, et al. Serum concentrations of polybrominated diphenyl ethers (PBDEs) and polybrominated biphenyl (PBB) in the United States population: 2003–2004. Environ Sci Technol. 2008;42(4):1377–1384. PubMed PMID: 18351120 [DOI] [PubMed] [Google Scholar]
- [56].Kafafi SA, Afeefy HY, Ali AH, et al. Binding of polychlorinated biphenyls to the aryl hydrocarbon receptor. Environ Health Perspect. 1993;101(5):422–428. Epub 1993/10/01. PubMed PMID: 8119253; PMCID: PMC1519849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Weisglas-Kuperus N, Patandin S, Berbers GA, et al. Immunologic effects of background exposure to polychlorinated biphenyls and dioxins in Dutch preschool children. Environ Health Perspect. 2000;108(12):1203–1207. Epub 2001/ 01/03. PubMed PMID: 11133402; PMCID: PMC1240203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Heilmann C, Budtz-Jorgensen E, Nielsen F, et al. Serum concentrations of antibodies against vaccine toxoids in children exposed perinatally to immunotoxicants. Environ Health Perspect. 2010;118(10):1434–1438. Epub 2010/06/22. PubMed PMID: 20562056; PMCID: PMC2957925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Park HY, Hertz-Picciotto I, Petrik J, et al. Prenatal PCB exposure and thymus size at birth in neonates in Eastern Slovakia. Environ Health Perspect. 2008;116(1):104–109. Epub 2008/01/17. PubMed PMID: 18197307; PMCID: PMC2199273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Terrell ML, Rosenblatt KA, Wirth J, et al. Breast cancer among women in Michigan following exposure to brominated flame retardants. Occup Environ Med. 2016;73(8):564–567. PubMed PMID: 27312402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Blanck HM, Marcus M, Tolbert PE, et al. Age at menarche and tanner stage in girls exposed in utero and postnatally to polybrominated biphenyl. Epidemiology. 2000;11(6):641–647. PubMed PMID: 11055623 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


