Abstract
Pregnane X receptor (PXR) is a master xenobiotic-sensing transcription factor and a validated target for immune and inflammatory diseases. The identification of chemical probes to investigate the therapeutic relevance of the receptor is still highly desired. In fact, currently available PXR ligands are not highly selective and can exhibit toxicity and/or potential off-target effects. In this study, we have identified garcinoic acid as a selective and efficient PXR agonist. The properties of this natural molecule as a specific PXR agonist were demonstrated by the screening on a panel of nuclear receptors, the assessment of the physical and thermodynamic binding affinity, and the determination of the PXR-garcinoic acid complex crystal structure. Cytotoxicity, transcriptional, and functional properties were investigated in human liver cells, and compound activity and target engagement were confirmed in vivo in mouse liver and gut tissue. In conclusion, garcinoic acid is a selective natural agonist of PXR and a promising lead compound toward the development of new PXR-regulating modulators.
1. Introduction
Pregnane X receptor (PXR or NR subfamily 1, group I, member 2, NR1I2) is universally recognized as a master regulator of key xenobiotic and drug metabolizing genes,1,2 such as the cytochrome P450 isoform 3A4 (CYP3A4), several phase II genes, and the multidrug resistance protein 1 (MDR1).3,4 Cloned in 1998 by two independent groups,5,6 the PXR gene is principally expressed in the liver and intestine.7,8 Its activation can be elicited by a number of ligands,3,4 including endogenous steroids, such as lithocholic acid and many other cholesterol metabolites, several classes of drugs and natural products, such as antibiotics, anticancer agents, the antihypertensive nifedipine, and the antifungal clotrimazole, herbal medicines such as the antidepressant hyperforin from St. John’s wort,9 the gut microflora products indole 3-propionic acid,10,11 and some dietary phytosterols and phenolics that include vitamin E.12,13 At the same time, a limited number of PXR antagonists and activity inhibitors have also been identified,4,14 including the drugs ketoconazole and A-792611, and the natural compounds sulforaphane, sesamine, coumestrol, and camptothecin, some of which have been investigated as drug resistance factors.
PXR activity is under the influence of molecular interactions with other nuclear receptors (NRs) and transcription factors that ultimately affect important physiological processes of the liver and small intestine. These include the regulation of inflammatory and metabolic pathways and the preservation of intestinal wall integrity.15−17
Over the years, intense research has been devoted to investigate the potential of PXR as a pharmacological target of human pathologies. The main ones include cholestatic liver disease,18 inflammatory bowel disease (IBD),19,20 and dyslipidemia.21 However, ligand promiscuity is a major obstacle in the pharmacological approach to such NR because most of the PXR modulators identified so far have other primary targets, and PXR itself could be considered an off-target in drug development.22
Studies on the antibiotic rifaximin (Xifaxan),19 a PXR agonist also used to treat diarrhea23 and hepatic encephalopathy,24 demonstrated the efficacy of this drug in the treatment of irritable bowel syndrome (IBS). Such therapeutic application received FDA approval in May 2015,25 and further studies demonstrated effects that were independent of the antibiotic activity of this compound on gut microbiota.26 As a consequence, PXR has emerged as a molecular target for the treatment of this syndrome.
Together these aspects indicate the need for the identification of novel and selective PXR agonists.
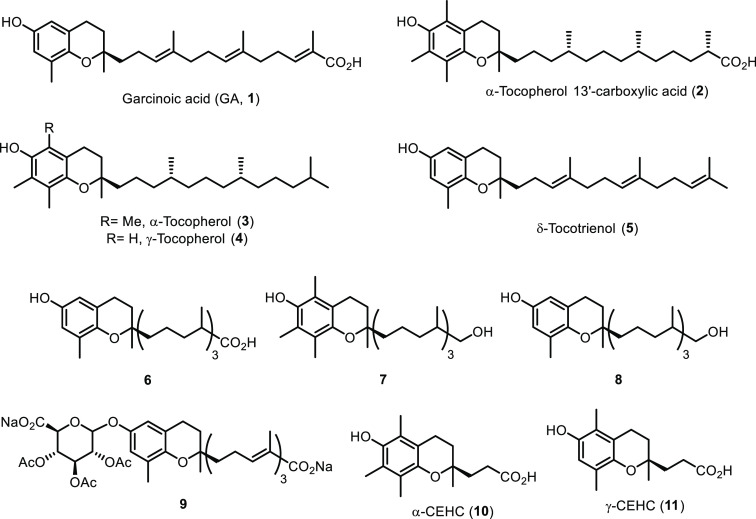
Recent studies suggested that some forms and metabolites of vitamin E might fulfill this aim. Podszun et al.27 demonstrated in LS 180 human colorectal adenocarcinoma cells that α-tocopherol 13′-carboxylic acid (2), a long-chain metabolite (LCM) of α-tocopherol (3),28,29 and δ-tocotrienol (5) (Chart 1)30,31 show PXR agonist function up-regulating the PXR-dependent gene p-glycoprotein. Other studies by some of us32 demonstrated an increased PXR expression in mouse brain and in vitro in embryonal cortical astrocytes treated with a natural analogue, garcinoic acid (GA, 1) (Chart 1), first described as a bioactive compound by Mazzini and colleagues.33 This is a plant derivative of δ-tocotrienol (5) with different African ethno-medicine applications.33,34 With this premise, herein we aim to explore the activity of GA (1) as PXR agonist. In pursuit of this aim, we isolated of GA (1) from Garcinia kola seeds and use the pure compound for metabolite synthesis and characterization (Chart 1). Most importantly, binding assays, gene expression profile, cocrystallization, and in vivo experiments were carried out to establish the efficacy of GA (1) as a PXR agonist and target engagement.
Chart 1. Chemical Structures of Garcinoic Acid (GA, 1), α- and γ-Tocopherol (3, 4), δ-Tocotrienol (5), Long-Chain Metabolites (2, 6–9), and Short-Chain Carboxyethylhydroxychroman (CEHC) Metabolites (10, 11).
2. Results
2.1. Isolation of Garcinoic Acid and Metabolite Synthesis
To isolate ≥95% pure GA (1) in sufficient amount for compound characterization and metabolite synthesis, the extraction procedures from Garcinia kola seeds available in the literature have been revised and optimized. Current extraction methods34,35 are based on Terashima protocol36 and consist of the alcoholic extraction of the seed followed by silica gel chromatography. Using our optimized approach, we were able to increase the purified GA (1) on the multigram scale. Three different parameters have been investigated to improve yield of extraction: seed mass/solvent ratio (1:1 or 2:1, w/v), temperature (25, 40, and 60 °C), and solvent (MeOH or EtOH) (see Table S2). The best conditions were obtained using MeOH as the solvent, at 25 °C and with a seeds/solvent ratio 1:1, w/v (entry 3, Table S2). Using the optimized protocol, 1.2 kg of finely ground G. kola seeds was extracted, affording 9.4 g of garcinoic acid (1) (0.78% yield w/w).
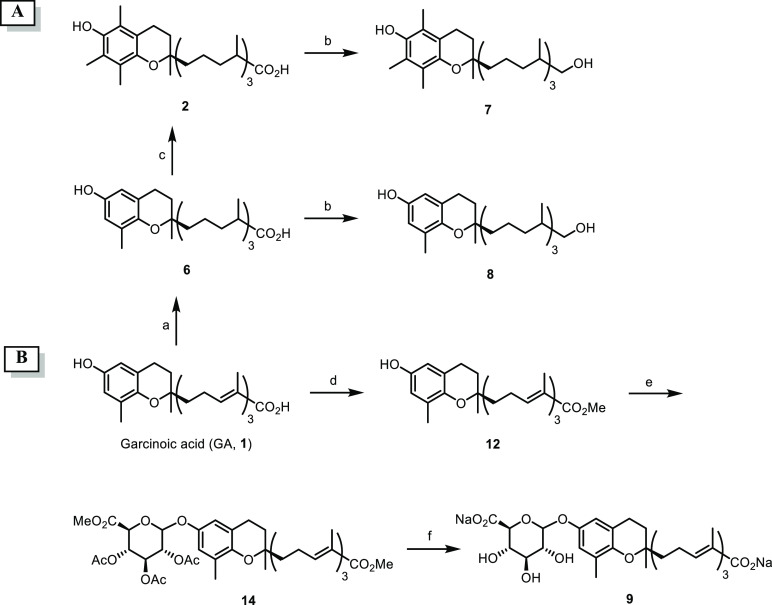
The synthesis of metabolites 2, 6–9 was performed according to Scheme 1A. In particular, hydrogenation of GA (1) was carried out under flow conditions using a packed Pd/C cartridge, afforded compound 6 in quantitative yield. Next, reduction of the carboxylic group by means of LiAlH4 in anhydrous THF at rt gave 13′-hydroxy-δ-tocopherol (8) in 87% isolated yield. 13′-Carboxy-δ-tocopherol (6) was also reacted with (CH2O)n in the presence of SnCl2 and HCl 12 M in Et2O at 70 °C to obtain α-tocopherol 13′-carboxylic acid (2) (92% yield) readily reduced to the corresponding alcohol derivative 7 with LiAlH4 in anhydrous THF (62% yield).33 The glucuronyl metabolite of GA (9) was prepared in a three-step synthetic sequence (Scheme 1B). Thus, GA ester (12), prepared by reacting 1 with CH2N2 in Et2O at −78 °C (89%), was coupled with methyl-1-bromo-2,3,4-tri-O-acetyl-α-d-glucuronate (13) and Fetizon reagent in toluene to give 14 in 19% overall yield after silica gel purification.37 Finally, mild basic hydrolysis (Na2CO3/MeOH, rt) provided the desired glucuronide 9 in 17% isolated yield over three steps after flash chromatography.
Scheme 1. Synthesis of Metabolites 2, 6–9.
Reagents and conditions: (a) 10% Pd/C cartridge (s-cart, 30 × 4 mm i.d.), 1 bar (full H2 mode), 25 °C, 1 mL min–1; (b) LiAlH4, THF, 0 °C → rt; (c) SnCl2, HCl 12 M, Et2O, (CH2O)n, 70 °C; (d) CH2N2, Et2O, 0 °C → rt; (e) methyl-1-bromo-2,3,4-tri-O-acetyl-α-d-glucuronate (13), Fetizon reagent, molecular sieves, toluene; (f) Na2CO3, MeOH, rt.
2.2. Identification of Garcinoic Acid as a Selective PXR Agonist
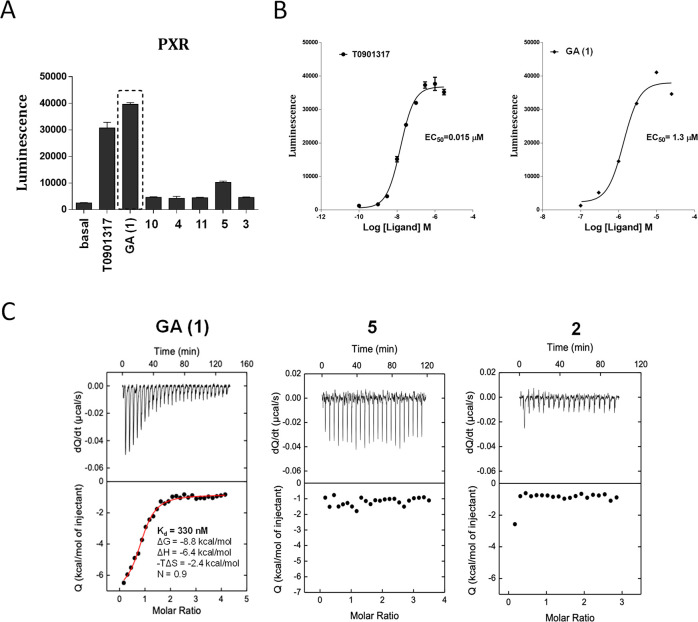
Ligand binding activity of GA (1) was explored via AlphaScreen technology over a panel of NRs that, along with hPXR (Figure 1A), included major human endocrine and metabolic receptors, such as the retinoid X receptor (RXR), constitutive androstane receptor (CAR), farnesoid X receptor (FXR), vitamin D receptor (VDR), liver X receptor (LXR) forms α and β, and the PPAR isoforms α, δ, and γ (see Figure S1). GA (1) was compared to its analogue δ-tocotrienol (5), the vitamers 3 and 4, their short-chain metabolites (SCMs) α-CEHC (10) and γ-CEHC (11), and the LCMs 2 and 8 (Chart 1). The test was performed in the presence of reference standard agonists for the different NRs. Among the vitamin E compounds, only GA showed significant PXR agonist activity (see Figure S1A). Much lower was the response of δ-tocotrienol (5), while all of the other vitamin E compounds were almost completely inactive at the receptor. Most importantly, GA (1) showed high selectivity for PXR, with only a low activity at the LXRβ receptor (see Figure S1B). None of the other vitamin E compounds showed any appreciable agonist activity on this receptor.
Figure 1.
PXR agonist activity and binding properties of GA. Binding activity of GA (1) was assessed by the AlphaScreen test at 10 μM compound concentration (A) and during a dose-dependent experiment (B). Comparisons were made with a series of vitamin E compounds and the PXR agonist 0.05 μM T0901317 (see Table S1). Calorimetric titrations of GA binding to PXR-LBD (C) were investigated in comparison with compounds 2 and 5 dissolved in DMSO and then further diluted to 60 μM final concentration in 25 mM Hepes buffer, pH 7.5, containing 150 mM NaCl. The final concentration of PXR-LBD in the reaction mixture was 3 μM, and DMSO was 8% v/v.
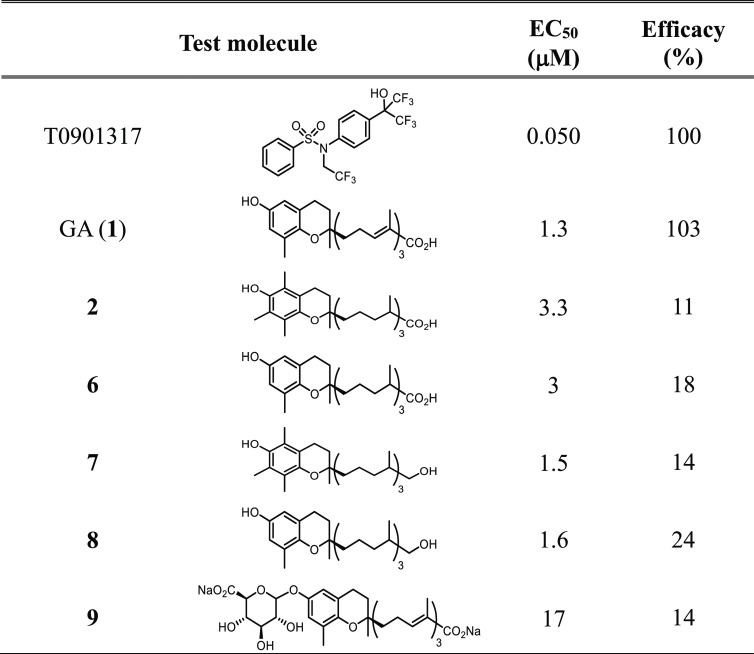
The activity of GA as PXR agonist was then evaluated in dose–response experiments in comparison with T090131738 (Figure 2B); the resulting EC50 values were 1.3 and 0.015 μM, respectively. Comparisons were also made with physiological LCM analogues 2, 6–9 (Table 1).
Figure 2.
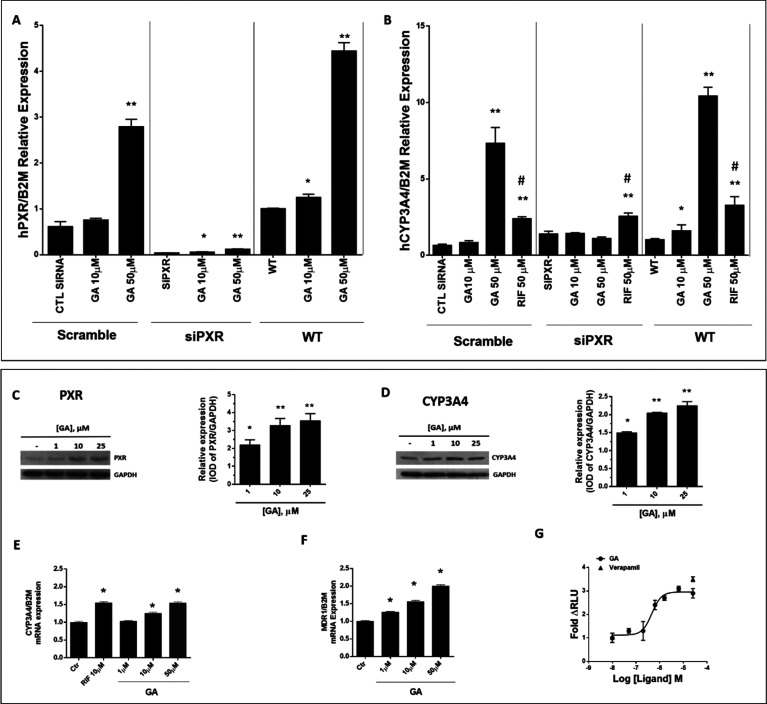
PXR and CYP3A4 expression in HepG2 cells treated with garcinoic acid (1). siRNA technique was used to transiently inhibit PXR (A) or CYP3A4 (B) gene expression [*p < 0.05; **p < 0.01 vs WT or Ctr test; #p < 0.01 vs Rifampicin (RIF)]. PXR (C) and CYP3A4 (D) protein and mRNA (E) expression were also assessed in cells treated for 24 h with 1 and 25 μM GA (1). MDR1 mRNA expression (F) was evaluated at concentrations between 1 and 50 μM GA. The P-glycoprotein activity (G) was measured in the presence of GA between 50 nM and 25 μM (●), and Verapamil (▲) was used as a control. t test: control versus treatments, *p < 0.05; **p < 0.01.
Table 1. Activity of GA (1) and Metabolites 2, 6–9 on PXR as Assessed by the AlphaScreen Testa.
Data represent mean values ± SDs of at least three independent experiments.
All of these compounds showed EC50 values similar to that of GA (1) (between 1.5 and 3.3 μM) with the exception of the glucuronide 9 that showed an EC50 value of 17 μM, suggesting an interfering role of phase II derivatization for GA agonist activity.39 The percentage of efficacy as compared to T0901317 was 103% for GA and ≤24% for the other LCMs (Table 1); worthy of note, the δ-configuration of the chromanol ring was associated with a higher agonist efficacy as compared to the α-configuration.
The specificity of GA agonist activity for PXR was demonstrated by the siRNA technique in HepG2 cells. When the PXR gene was transiently inhibited, a reduction of both the PXR and the CYP3A4 gene transcription responses to GA treatment was confirmed (Figure 2A and B, respectively).
2.3. Calorimetric Titration of Garcinoic Acid Binding to the PXR Ligand Binding Domain
To obtain direct proof of interaction with the hPXR ligand binding domain (LBD), we performed an isothermic titration calorimetry (ITC) experiment using His-tagged hPXR LBD protein in solution (Figure 1C). ITC measures the affinity, Ka, the Gibbs energy (ΔG = −RT ln Ka), and the changes in enthalpy, ΔH, and entropy, ΔS, associated with the binding of the FKK compounds (ΔG = −RT ln K = ΔH – TΔS). Enthalpic and entropic contributions to binding affinity define the nature of the forces that drive the binding reaction.40,41 GA (1) binds to a single site in LBD (Kd = 330 nM, ΔG = −8.8 kcal/mol, ΔH = −6.4 kcal/mol, −TΔS = −2.4 kcal/mol, N = 0.9) (Figure 1C, left panel). In contrast, the other PXR inactive analogues, δ-tocotrienol (5) and α-tocopherol 13′-carboxylic acid (2), do not show any detectable binding to PXR LBD in this assay (Figure 1C, middle and right panels, respectively).
2.4. Crystal Structure of PXR–Garcinoic Acid Complex
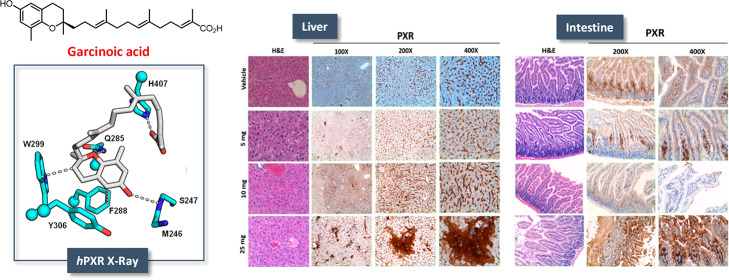
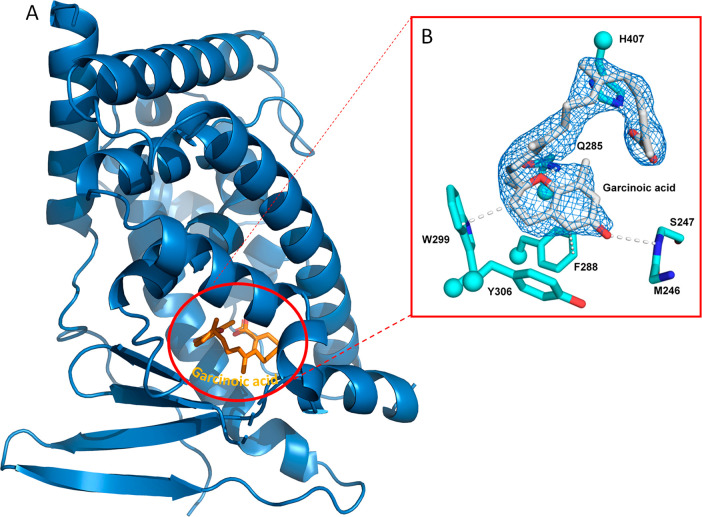
To determine the structural basis of the GA–PXR binding interaction, crystals of the LBD of hPXR incubated with GA (1) were grown successfully. The resultant crystals diffracted X-rays to 2.3 Å resolution and revealed GA bound in a single orientation within the PXR ligand binding pocket of LBD (Figure 3 and Table S2). The fused ring moiety of GA contacts a set of three aromatic residues (F288, W299, and Y306) in the PXR LBD, forming an array of van der Waals contacts including face-to-face (F288) and edge-to-face (W299) π–π interactions. Other specific contacts include hydrogen bonds with the endocyclic oxygen and phenolic hydroxyl of GA with Q285 and the backbone of S247, respectively (Figure 3). Last, the carboxylate moiety of GA (1) forms an ionic interaction with H407. Taken together, the hPXR LBD specifically recognizes GA with numerous contacts that corroborate the potent binding of this ligand.
Figure 3.
Crystal structure of hPXR–garcinoic acid complex. (A) Overview of hPXR LBD–garcinoic acid complex. (B) 2.3 Å resolution X-ray diffraction data of crystals demonstrated that GA (gray) binds in a single orientation within the ligand binding pocket of hPXR, contacted by four amino acid side chains (cyan) and one main-chain region (M246–S247; cyan). Distances noted are in angstroms.
2.5. In Vitro and In Vivo Target Engagement
In Vitro Data
To confirm the activity of GA as a PXR agonist, PXR protein expression was investigated in human hepatocarcinoma cells (HepG2) and immortalized hepatic progenitor cells (HepaRG) that were preliminarily investigated for compound toxicity (see Figure S2).
In HepG2 cells, GA increased in a dose–dose-dependent manner the PXR protein expression (Figure 2C), the CYP3A4 protein and mRNA expression (Figure 2D and E, respectively), and the MDR1 gene expression and activity (Figure 2F and G, respectively). Even higher was the response of PXR protein to GA treatment in HepaRG cells (see Figure S3A). This activity of GA was significantly higher as compared to that of the α-TOH metabolites 2 and 7 (see Figure S3A). Moreover, GA reverted the PXR binding and antagonist effect of sulforaphane (see Figure S3C).42
The GA-induced stimulation of PXR protein expression was investigated for its functional modulation on the CYP450-mediated ω-oxidation and subsequent catabolism of the α-TOH side chain.30 In HepG2 cells, GA treatment did not modify the level of α-tocopherol (3) that was rapidly taken up, compensating the depletion of this vitamin observed under standard cell culture conditions (see Figure S4A).43 On the contrary, GA stimulated both the CYP450-mediated production and the efflux of α-tocopherol metabolites (see Figure S4A–C). The latter finding confirms the role of GA as an activator of the PXR-dependent membrane transporter MDR1 shown earlier in Figure 2F and G, and as was already described in the literature for other LCMs, such as 2.27 GA was found to increase PXR and CYP3A4 protein expression also in the human colorectal cancer cell line HT29 and in its mucous-secreting counterpart HT29-MTX (Figure S6A–D).
In Vivo Data
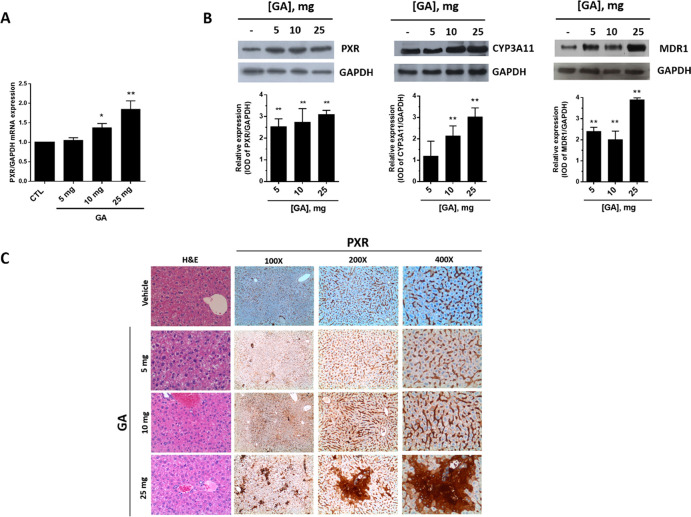
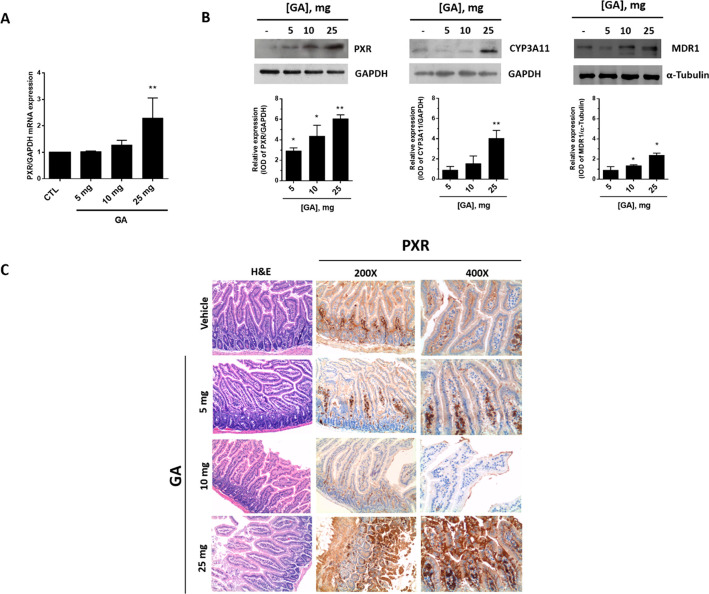
GA toxicity and PXR activation were investigated in mice after acute (single bolus) administration. At observation (24 h after the treatment), mortality was 100% in the group treated with 100 mg of GA and 33% in the group that received 50 mg of GA (1). Doses of ≤25 mg were well tolerated as suggested by the gross appearance and behavior of the treated animals and by the post-mortem examination of liver tissue and all main organs that revealed the complete absence of signs of damage. A dose-dependent stimulation effect of GA on PXR protein and mRNA was observed in liver tissue (Figure 4A and B, respectively). PXR mRNA also increased in intestine upon GA treatment (Figure 5A). Such transcriptional response, however, was not sufficient to significantly increase PXR protein expression in this tissue (not shown), possibly due to the lower PXR levels expressed in this tissue in comparison with liver tissue (Figure 4A). GA also increased CYP3A4 (which corresponds to CYP3A11 isoform in mice) protein expression in both of the two tissues (Figures 4B and 5B).
Figure 4.
PXR, CYP3A4, and MDR1 expression in the liver of mice treated with garcinoic acid (1). Mice were treated with increasing doses of GA from 5 to 25 mg administered as a single bolus, and PXR mRNA (A) and protein expression (B, left panel) were measured in liver samples 24 h post-treatment. CYP3A4 (CYP3A11 in mice) and MDR1 protein expression were also investigated (B, middle and right panels); *p < 0.05; **p < 0.01. Liver histology was examined by hematoxylin and eosin (H&E) staining and IHC for PXR antigen (C).
Figure 5.
PXR, CYP3A4, and MDR1 expression in the intestine of mice treated with garcinoic acid (1). Mice were treated with increasing doses of GA from 5 to 25 mg administered as a single bolus, and PXR mRNA (A) and protein expression (B, left panel) were measured in liver samples 24 h post-treatment. CYP3A4 (CYP3A11 in mice) and MDR1 protein expression were also investigated (B, middle and right panels); *p < 0.05; **p < 0.01. Liver histology was examined by hematoxylin and eosin (H&E) staining and IHC for PXR antigen (C).
The response of PXR protein to GA treatment in liver and gut was confirmed by IHC evaluation. In the hepatic parenchyma (Figure 4C), PXR was mainly localized in sinusoidal endothelial cells. In mice treated with 25 mg of GA, PXR staining also localized in hepatocytes, particularly around centrolobular veins (zone 3) and the lobular midzonal area (zone 2). In small bowel from control group mice, PXR expression localized exclusively in the lamina propria, lymphocytes, and plasma cells of intestinal villi, without any significant staining of enterocytes (Figure 5C). On the contrary, in mice treated with GA, a strong and diffuse PXR expression was observed in villous epithelium (enterocytes) and focally also in the glandular epithelium of crypts at the base of mucosa. Furthermore, a weak staining was present on the brush border of enterocytes in mice treated with 10 mg of GA (Figure 5C).
3. Discussion and Conclusions
The present study conclusively demonstrates that GA (1) is an efficient natural agonist of PXR. This finding was obtained by optimizing the isolation protocol of 1 to reach a level of purity and quantity of the compound sufficient to carry out the synthesis of other metabolites and to get insight into compound binding and activity at PXR. These properties of GA were investigated with different approaches, including screening on a panel of NRs, physical and thermodynamic (calorimetric) evaluation of binding affinity, cocrystallization experiments, and then in vitro and in vivo transcriptional and functional assessment.
In line with the in vitro binding affinity results, crystallographic data were indicative of GA binding potency that can be explained by the presence of several and stable interactions (Figure 3). These include van der Waals contacts between the chroman ring of GA and three aromatic residues (F288, W299, and Y306), hydrogen bonds established by the endocyclic oxygen and phenolic hydroxyl group of GA with residue Q285 and the backbone of S247, and the crucial ionic interaction of the carboxylate moiety of GA with H407 (Figure 4).
The transcriptional function of PXR affects genes encoding drug-metabolizing enzymes and transporters to essentially detoxify and eliminate xenobiotics and endotoxins.1,2,15 This gene regulation function of PXR has been explained on the basis of its interaction with other NRs and regulatory proteins; upon activation, PXR typically heterodimerizes with the retinoid X receptor (RXR) to recruit coactivators, instead of corepressors, that modulate the binding of the heterodimer with specific responsive elements in the promoter regions of target genes. For example, in the proximal promoter region of CYP3A4, a repeat of one-half hexamers divided by six nucleotides was recognized as a proximal PXR responsive element (2169/2152).6 Additionally, the CYP3A4 promoter includes the xenobiotic responsive enhancer module (XREM), a distal enhancer sequence containing two extra PXR responsive elements.44−46
MDR1 (or P-gp) is also a PXR reporter gene in liver cells, and recent reports showed the hepatic metabolite 2 as able to stimulate this gene,27 consistent with a role of PXR not only as xenosensor15 and master regulator of vitamin E metabolism,48 but also as a sensor of its biotransformation products28 and candidate receptor for this family of natural products with marked selectivity for dimethyl and unsaturated forms.
In this respect, GA shows unique properties among the entire family of vitamin E compounds investigated in this study, being the most potent enhancer of PXR activity and consequently of vitamin E biotransformation in human liver cells. In fact, the upregulation of key genes in vitamin E metabolism, such as CYP3A430,31 and the phase III gene MDR1,27,29 stimulates the ω-hydroxylation of cellular α-tocopherol (3) and the efflux of its LCMs 7 and 2 (Figure S3) that definitely are bioactive metabolites.27,29,49 These aspects also suggest that PXR may play a central role in the GI tract, particularly in the liver, to discriminate between α-tocopherol, other vitamers, and metabolites of vitamin E.13,28,29
Important enough, after we screened the binding of GA over a panel of NRs, this compound showed selective agonist activity at PXR (see Figure S1). No response to GA and other analogues with tocol-like structure was observed on PPARs and FXR, RAR, and RXR. CAR, another NR originally proposed to help explain the CYP3A-dependent metabolism of vitamin E in human hepatocytes30,48 and characterized by constitutive activation, was partially inhibited during affinity binding experiments with GA. These findings support selective action of GA for PXR and shed light on future applications of GA in targeted chemoprevention and therapy protocols of PXR-expressing organs, such as the liver, intestine, and to a slighter extent the kidney. In fact, the role of PXR as transcriptional regulator of xenobiotic and drug metabolizing genes with a broad substrate promiscuity3 has represented the major drawback for the pharmacological approach to the receptor.22 In this respect, GA represents a novel and selective PXR agonist, holding great potential in the development of therapeutic agents for a range of human pathologies that include cholestatic liver disease,18 dyslipidemia,21 and IBD;19,20 furthermore, recently rifaximin has received FDA approval for IBS treatment.25
Our in vivo data unequivocally demonstrate that both intestinal and liver PXR respond to the agonist activity of GA, thus providing mechanistic support for the investigation of this natural compound in PXR-related diseases of the liver and gut. In a recent study, we also demonstrated PXR agonist activity of GA in the mouse brain as well as in isolated astrocytes,32 compatible with the role of this NR in the ApoE-mediated detoxification of amyloid-β peptide, a molecule with a pathogenic role in Alzheimer’s disease.
The present findings on PXR agonist activity of GA add further and important information to the biological activity of this natural analogue of vitamin E.34 In this context, GA can be exploited as a valuable chemical probe and lead compound for medicinal chemistry explorations to disclose novel therapeutic agents for the prevention and treatment of lipotoxicity and chronic inflammatory diseases, such as atherosclerosis,34 IBD, nonalcoholic fatty liver disease, and steatohepatitis.50,51 Current studies are directed toward the preparation of GA (1) derivatives with improved potency, molecular properties, and metabolic stability, the results of which will be reported in due course.
4. Experimental Section
4.1. Chemistry
General Methods
Unless otherwise noted, chemicals were obtained from commercial suppliers and used without further purification. NMR spectra were recorded on a Bruker AC 400 MHz spectrometer in the indicated solvent. Chemical shifts are reported in parts per million (ppm) and are relative to CDCl3 (7.26 and 77.0 ppm) or to CD3OD (3.31 and 49.2 ppm). The abbreviations used are as follows: s, singlet; brs, broad singlet; d, doublet; dd, double doublet; t, triplet; q, quartet; m, multiplet; brm, broad multiplet. Melting points were determined by the capillary method using a Buchi 535 instrument, and they were not corrected. TLC was performed on aluminum backed silica plates (silica gel 60 F254). Flash chromatographic purifications were performed via Biotage Isolera Prime using the appropriate cartridge, eluent, and gradient. Hydrogenations were performed with H-Cube apparatus (Thalesnano Nanotechnology Inc., Budapest, Hungary) using a 10% Pd/C cartridge (s-cart, 30 × 4 mm i.d.). With the exception of compound 9, all of the synthesized compounds have been previously reported, and their spectroscopic and analytical data were consistent with the literature.33,35,52 The purity of the synthesized compounds (>95%) was assessed by HPLC–HRMS.
Isolation of Garcinoic Acid (1)
Finely ground seeds (1.2 kg) of Garcinia kola were suspended in MeOH (1.2 L) and stirred at 25 °C for 6.5 h. The resulting suspensions were filtered off under vacuum, and the resulting solid residue was resuspended in MeOH (1.2 L) and stirred at 25 °C for a further 17.5 h. The suspension was filtered again under vacuum, affording a crude brownish oil (56 g). The crude was purified by silica gel flash chromatography (eluent: CH2Cl2/MeOH, from 100:0 to 90:10, v/v), affording 9.4 g (yield 0.78%, w/w) of ≥95% pure (determined by HPLC–HRMS analysis) garcinoic acid (1) as a yellow-green oil.521H NMR (400 MHz, CDCl3): δ 1.27 (3H, s), 1.53–1.61 (1H, m), 1.63 (6H, m), 1.75–1.79 (2H, m), 1.84 (3H, s), 1.99–2.01 (2H, m), 2.06–2.13 (9H, m), 2.20 (3H, s), 2.29–2.30 (2H, m), 2.70 (2H, t, J = 6.49 Hz), 5.13–5.14 (2H, m), 6.41 (1H, d, J = 2.51 Hz), 6.50 (1H, d, J =2.52 Hz), 6.88–6.92 (1H, m). 13C NMR (100.6 MHz, CDCl3): δ 12.0, 15.8, 15.9, 16.0, 22.1, 22.4, 24.0, 26.4, 27.5, 31.3, 38.0, 39.5, 42.0, 77.3, 112.6, 115.7, 121.2, 124.4, 125.1, 126.9, 127.3, 133.7, 134.8, 145.0, 145.9, 147.7, 173.3.
Crystallization, X-ray Diffraction Data Collection, and Structure Refinement
PXR-LBD-SRC-1p was incubated with 1 mM GA and then crystallized in 30% (v/v) MPD and 100 mM imidazole/HCl pH 7.0 by the hanging drop vapor diffusion method at 4 °C. Diffraction data were collected at 100 K at APS Beamline 23-ID-D. Data were reduced in XDS54,55 and scaled in Aimless.55 The tethered PXR-LBD/SRC-1p-GA structure was solved via molecular replacement in Phenix using the tethered PXR-LBD/SRC-1p apo structure (PDB: 3CTB). The model was refined using Phenix with manual adjustments using Coot as necessary. Coordinates and structure factors have been deposited to the PDB with accession code 6P2B.
4.2. Biology
Cloning, Expression, and Purification of PXR-LBD/SRC-1p
Tethered PXR-LBD/SRC-1p was synthesized via GenScript. The gene product was subsequently subcloned into the pLIC-His pMCSG7 expression vector. Site-directed mutagenesis of the N-terminal tag was performed to replicate the construct used for a previously determined PXR crystal structure (PDB: 3CTB).53 The final expression vector containing the His-tagged PXR-LBD/SRC-1p construct was transformed into E. coli BL21(DE3) cells. A 100 mL overnight culture supplemented with 100 μg/mL ampicillin was used to inoculate a 1.5 L culture supplemented with 100 μg/mL ampicillin at 37 °C until the OD600 was approximately 0.6. The temperature was reduced to 18 °C, and expression was induced with the addition of 0.1 mM isopropyl β-d-thiogalactopyranoside and continued overnight. The next day, cells were harvested and frozen at −80 °C prior to purification. Protein was purified by Ni affinity chromatography and size exclusion chromatography using the following buffers: Ni A – 25 mM HEPES, pH 7.9, 5% v/v glycerol, 150 mM NaCl, 1 mM DTT, 10 mM imidazole; Ni B – 25 mM HEPES, pH 7.9, 5% v/v glycerol, 150 mM NaCl, 1 mM DTT, 250 mM imidazole; GF – 25 mM HEPES, pH 7.9, 5% v/v glycerol, 150 mM NaCl, 5 mM DTT.
AlphaScreen Assay of Nuclear Receptor Binding
The activity of nuclear receptors was determined using AlphaScreen technology, a recruitment coactivator assay previously described.56
Expression and Purification of His-Tagged PXR Ligand Binding Domain (LBD) Protein
The expression and purification of His-tagged PXR Ligand Binding Domain (LBD) protein were performed as published with minor modifications.57 For the expression of the protein, Luria–Bertani (LB) media were inoculated with a saturated culture of BL21-Gold cells transformed with HIS-LIC plasmid containing the PXR LBD construct. The mixture was then allowed to shake at 37 °C until the cells reached an OD600 ≈ 0.6. The temperature was then reduced to 18 °C, at which time IPTG was added (final concentration of 0.1 mM) to induce protein expression. For purification of PXR-LBD, the His-tag was not removed, and the uncleaved protein was loaded onto the gel filtration column with buffer [HEPES (25 mM, pH 7.5) and NaCl (150 mM)].
Isothermal Titration Calorimetry (ITC)
ITC was performed using a VP-ITC microcalorimeter from MicroCal/Malvern Instruments (Northampton, MA). The protein and the ligands (in DMSO, 8% vol/vol final concentration) were prepared in 25 mM Hepes, pH 7.5 with 150 mM NaCl. In all of the experiments, the ligand solution was injected in 10-μL aliquots into the calorimetric cell containing PXR-LBD [3 μM] at 37 °C. The respective concentrations of GA (1), 2, and 5 in the syringe were 60 μM each. The heat evolved upon each injection of the ligands was obtained from the integral of the calorimetric signal. The heat associated with binding to PXR-LBD in the cell was obtained by subtracting the heat of dilution from the heat of reaction. The individual heats were plotted against the molar ratio, and the enthalpy change (ΔH), association constant (Ka = 1/Kd), and stoichiometry were obtained by nonlinear regression of the data.
Cell Studies
HepG2 (human hepatoma cell line; ATCC HB-8065, Manassas, VA) and HepaRG (HPRGC10, Thermo Fisher Scientific) cells were maintained in culture as previously described.47 Experiments were performed between passages 2 and 10. Cell viability, clonogenic activity, and apoptotic cell death were assessed as previously described (see Figure S2).58,59 HT29 human colorectal adenocarcinoma cells (ATCC HTB-38) and its mucous-secreting HT29-MTX subclone (Sigma-Aldrich) were used as intestinal cell models. HT29 cells were cultured in RPMI (Invitrogen; Life Technologies) containing 10% FBS (v/v) and 1% (v/v) l-glutamine. HT29-MTX cells (from Sigma-Aldrich) were cultured in DMEM supplemented with 2 mM glutamine, 1% nonessential amino acids, and 10% FBS. Cells were maintained at 37 C in a humidified 5% (v/v) CO2 atmosphere chamber and were used between passages 10 and 20.
Immunoblot
Cells were harvested and lysed in ice-cold cell lysis buffer (Cell Signaling Technology) and 20 μL/mL protease inhibitor cocktail (Roche). Murine tissues (10 mg each) were weighed and resuspended in 300 μL of PBS supplemented with NP40 (1:50) and a cocktail of protease inhibitors (Pierce, Thermo Fisher Scientific). After preliminary homogenation in a 1-mL potter, sonication was performed in ice (3 cycles of 15 s each with 1 min intervals). After incubation in ice for 1 h and centrifugation (12 000 rpm for 20 min at 4 °C), the supernatant was recovered, and total proteins were quantified by the BCA assay kit (Pierce, Thermo Fisher Scientific). Twenty micrograms of proteins was resolved by 10–12% SDS-PAGE, and immunoblot was performed using anti-PXR (H-160, 1:200 dilution; sc-25381, Santa Cruz Biotechnology, Inc.), anti-PXR (1:1000 dilution, ab192579, abcam), anti-CYP3A4 (1:1000 dilution; TA324142, OriGene Technologies, Inc., Rockville, MD), anti-GAPDH (D16H11, 1:1000 dilution; #5174, Cell Signaling Technology, Beverly, MA), and anti-MDR1 (dilution 1:1000, (D3H1Q) Rabbit mAb #12683 Cell Signaling Technology, Beverly, MA, and alpha-tubulin (dilution 1:1000, Cell Signaling Technology, Beverly, MA) as primary antibodies. A horseradish peroxidase–conjugated antirabbit IgG (1:2000 dilution; #7074, Cell Signaling Technology, Beverly, MA) was used as the secondary antibody. The band intensity was analyzed using Gel Pro analyzer Software. The results of immunoblots are shown as relative expression of the investigated protein by correction for housekeeping protein and control test (value = 1) in each series of experiments.
P-Glycoprotein Activity Assay
To evaluate the effects of GA on recombinant human Pgp in a cell membrane fraction, we used the Pgp-Glo assay system (Promega, Madison, WI), according to the manufacturer’s instruction. The assay relies on the ATP dependence of the light-generating reaction of firefly luciferase.
Gene Expression and siRNA PXR Transfection
RNA was extracted using the RNeasy Plus Kit (QIAGEN, Valencia, CA) and quantified with Implen NanoPhotometer (GmbH Germany), and cDNA synthesis was carried out using SuperScript IV (Thermo Fisher Scientific, MA). For siRNA PXR transfection, HepG2 cells were transfected with siRNAs PXR using Lipofectamine RNAiMAX Transfection Reagent (Thermo-Fischer). Reverse transfection was also carried out, and after the silencing procedure (24 h) HepG2 cells were treated with GA for a further 24 h.
In Vivo Studies
The 6–7-weeks old C57BL/6 wild-type male mice from Charles River were kept under standard environmental conditions (22 °C, 35% relative humidity, 12 h dark/light cycle) with free access to tap water and a standard diet. After being shipped, mice were allowed to adapt to the new environment before the experiments were initiated. All animal experiments were approved by the Institutional Animal Care and Use Committee of the University of Perugia as well as all surgeries and treatments. Animals were divided into six treatment groups (three animals each): control (vehicle = olive oil) and GA treated groups at the dosages of 5, 10, 25, 50, and 100 mg. GA was administered by oral gavage as a single bolus, and the animals were sacrificed after 24 h according to institutional guidelines to collect organs for histology and biochemistry evaluation as described in the other sections.
IHC Assay
Liver and gut mouse specimens were formalin-fixed and paraffin-embedded. Four micrometer sections were mounted on polarized glass slides and stained with hematoxylin and eosin (Sigma-Aldrich). Immunohistochemistry analysis of PXR (polyclonal antibody; dilution 1:200; ab217375, abcam) was performed using the Bond III (Vision BioSystems, Buffalo Grove, IL) automated slide preparation system, and Polymer Refine Detection (Vision BioSystems) was used as the antibody detection system. 3,3′-Diaminobenzidine (DAB) was used as chromogen. The section was then counterstained with hematoxylin (Sigma-Aldrich) and mounted with DPX (06522, Sigma-Aldrich).
Statistical Analysis
Statistically significant differences were determined using one-way analysis of variance followed by Tukey’s multiple comparison test as the post hoc test or t test, and differences were considered statistically significant at p < 0.05.
Glossary
Abbreviations Used
- CAR
constitutive androstane receptor
- FXR
farnesoid x receptor
- GA
garcinoic acid
- IBD
intestinal bowel disease
- IBS
irritable bowel syndrome
- ITC
isothermal titration calorimetry
- LBD
ligand binding domain
- LCM
long-chain metabolite
- LXR
liver x receptor
- NR
nuclear receptor
- MDR1
multidrug resistance protein 1
- PPAR
peroxisome proliferator-activated receptor
- PXR
pregnane x receptor
- RXR
retinoid x receptor
- SCM
short-chain metabolite
- α-TOH
alpha-tocopherol
- δ-T
gamma-tocotrienol
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jmedchem.0c00012.
Supplementary figures including general methods, synthetic procedures, analytical and spectroscopic characterization of compounds 2 and 5–8, NMR spectra and HPLC–HRMS copies, ligand binding activity of GA and other vitamin E analogues on a series of nuclear receptors, cell-base tests and in vivo assays, crystallographic data collection, and refinement statistics (PDF)
Molecular formula strings (CSV)
Author Contributions
● D.B. and F.D.F. contributed equally.
Part of this research was supported by the Italian Ministry of University grant program, the National Technology Agrifood Cluster, Health and Nutrition program, and the PROS.IT project (CTN01_00230_413096; completed). A.G. and F.G. have also been supported by the “Ricerca di base” grant program of the University of Perugia (completed). At the time of this research, D.B. was a postdoc fellow of the FIRC-AIRC young investigator’s grant program. S.M. was funded by NIH grants R01 CA127231 (completed), CA 161879 (completed), CA222469, Department of Defense Partnering PI (W81XWH-17-1-0479; PR160167), R01 ES030197, as well as R43DK105694 (PI: Jay Wrobel, completed) and P30DK041296 (PI: Alan Wolkoff) (pilot award completed). M.R.R was funded by NIH grants CA098468 and CA207416.
The authors declare the following competing financial interest(s): A.G. and R.P. are cofounders of TES Pharma.
Supplementary Material
References
- Kandel B. A.; Thomas M.; Winter S.; Seehofer D.; Burk O.; Schwab M.; Zanger U. M. Genomewide comparison of the inducible transcriptomes of nuclear receptors CAR, PXR and PPARalpha in primary human hepatocytes. Biochim. Biophys. Acta, Gene Regul. Mech. 2016, 1859, 1218–1227. 10.1016/j.bbagrm.2016.03.007. [DOI] [PubMed] [Google Scholar]
- Kliewer S. A.; Goodwin B.; Willson T. M. The nuclear pregnane X receptor: a key regulator of xenobiotic metabolism. Endocr. Rev. 2002, 23, 687–70. 10.1210/er.2001-0038. [DOI] [PubMed] [Google Scholar]
- Chen Y.; Tang Y.; Guo C.; Wang J.; Boral D.; Nie D. Nuclear receptors in the multidrug resistance through the regulation of drug-metabolizing enzymes and drug transporters. Biochem. Pharmacol. 2012, 83, 1112–1126. 10.1016/j.bcp.2012.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee M.; Chen T. Differential regulation of CYP3A4 promoter activity by a new class of natural product derivatives binding to pregnane X receptor. Biochem. Pharmacol. 2013, 86, 824–835. 10.1016/j.bcp.2013.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliewer S. A.; Moore J. T.; Wade L.; Staudinger J. L.; Watson M. A.; Jones S. A.; McKee D. D.; Oliver B. B.; Willson T. M.; Zetterström R. H.; Perlmann T.; Lehmann J. M. An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell 1998, 92, 73–82. 10.1016/S0092-8674(00)80900-9. [DOI] [PubMed] [Google Scholar]
- Blumberg B.; Sabbagh W. Jr.; Juguilon H.; Bolado J. Jr.; van Meter C. M.; Ong E. S.; Evans R. M. SXR, a novel steroid and xenobiotic-sensing nuclear receptor. Genes Dev. 1998, 12, 3195–3205. 10.1101/gad.12.20.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerberg L.; Hallström B. M.; Oksvold P.; Kampf C.; Djureinovic D.; Odeberg J.; Habuka M.; Tahmasebpoor S.; Danielsson A.; Edlund K.; Asplund A.; Sjöstedt E.; Lundberg E.; Szigyarto C. A.; Skogs M.; Takanen J. O.; Berling H.; Tegel H.; Mulder J.; Nilsson P.; Schwenk J. M.; Lindskog C.; Danielsson F.; Mardinoglu A.; Sivertsson A.; von Feilitzen K.; Forsberg M.; Zwahlen M.; Olsson I.; Navani S.; Huss M.; Nielsen J.; Ponten F.; Uhlén M. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteomics 2014, 13, 397–406. 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- https://www.ncbi.nlm.nih.gov/gene/8856#gene-expression (accessed Nov. 15, 2019).
- Hogle B. C.; Guan X.; Folan M. M.; Xie W. PXR as a mediator of herb-drug interaction. J. Food Drug Anal. 2018, 26, S26–S31. 10.1016/j.jfda.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikoff W. R.; Anfora A. T.; Liu J.; Schultz P. G.; Lesley S. A.; Peters E. C.; Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 3698–3703. 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh M.; Mukherjee S.; Wang H.; Li H.; Sun K.; Benechet A. P.; Qiu Z.; Maher L.; Redinbo M. R.; Phillips R. S.; Fleet J. C.; Kortagere S.; Mukherjee P.; Fasano A.; Le Ven J.; Nicholson J. K.; Dumas M. E.; Khanna K. M.; Mani S. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity 2014, 41, 296–310. 10.1016/j.immuni.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli F. Interactions of polyphenolic compounds with drug disposition and metabolism. Curr. Drug Metab. 2007, 8, 830–838. 10.2174/138920007782798180. [DOI] [PubMed] [Google Scholar]
- Galli F.; Azzi A.; Birringer M.; Cook-Mills J. M.; Eggersdorfer M.; Frank J.; Cruciani G.; Lorkowski S.; Özer N. K. Vitamin E: emerging aspects and new directions. Free Radical Biol. Med. 2017, 102, 16–36. 10.1016/j.freeradbiomed.2016.09.017. [DOI] [PubMed] [Google Scholar]
- Mani S.; Dou W.; Redinbo M. R. PXR antagonists and implication in drug metabolism. Drug Metab. Rev. 2013, 45, 60–72. 10.3109/03602532.2012.746363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oladimeji P. O.; Chen T. PXR: more than just a master xenobiotic receptor. Mol. Pharmacol. 2018, 93, 119–127. 10.1124/mol.117.110155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cave M. C.; Clair H. B.; Hardesty J. E.; Falkner K. C.; Feng W.; Clark B. J.; Sidey J.; Shi H.; Aqel B. A.; McClain C. J.; Prough R. A. Nuclear receptors and nonalcoholic fatty liver disease. Biochim. Biophys. Acta, Gene Regul. Mech. 2016, 1859, 1083–1099. 10.1016/j.bbagrm.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Velazquez J. A.; Carrillo-Cordova L. D.; Chavez-Tapia N. C.; Uribe M.; Mendez-Sanchez N. Nuclear receptors in nonalcoholic fatty liver disease. J. Lipids 2012, 2012, 139875. 10.1155/2012/139875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakizaki S.; Takizawa D.; Tojima H.; Horiguchi N.; Yamazaki Y.; Mori M. Nuclear receptors CAR and PXR; therapeutic targets for cholestatic liver disease. Front. Biosci., Landmark Ed. 2011, 16, 2988–3005. 10.2741/3893. [DOI] [PubMed] [Google Scholar]
- Cheng J.; Shah Y. M.; Gonzalez F. J. Pregnane X receptor as a target for treatment of inflammatory bowel disorders. Trends Pharmacol. Sci. 2012, 33, 323–330. 10.1016/j.tips.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah Y. M.; Ma X.; Morimura K.; Kim I.; Gonzalez F. J. Pregnane X receptor activation ameliorates DSS-induced inflammatory bowel disease via inhibition of NF-kappaB target gene expression. Am. J. Physiol. Gastrointest. Liver Physiol. 2007, 292, G1114–1122. 10.1152/ajpgi.00528.2006. [DOI] [PubMed] [Google Scholar]
- Gao J.; Xie W. Targeting xenobiotic receptors PXR and CAR for metabolic diseases. Trends Pharmacol. Sci. 2012, 33, 552–558. 10.1016/j.tips.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau A. J.; Yang G.; Yap C. W.; Chang T. K. Selective agonism of human pregnane X receptor by individual ginkgolides. Drug Metab. Dispos. 2012, 40, 1113–1121. 10.1124/dmd.112.045013. [DOI] [PubMed] [Google Scholar]
- DuPont H. L. Therapy for and prevention of traveler’s diarrhea. Clin. Infect. Dis. 2007, 45 (Suppl. 1), S78–84. 10.1086/518155. [DOI] [PubMed] [Google Scholar]
- Lawrence K. R.; Klee J. A. Rifaximin for the treatment of hepatic encephalopathy. Pharmacotherapy 2008, 28, 1019–1032. 10.1592/phco.28.8.1019. [DOI] [PubMed] [Google Scholar]
- Administration USFDA . FDA Approves Two Therapies to Treat IBS-D, Release FN; U.S. Food and Drug Administration: Silver Spring, MD, 2015. [Google Scholar]
- Sartor R. B. Review article: the potential mechanisms of action of rifaximin in the management of inflammatory bowel diseases. Aliment. Pharmacol. Ther. 2016, 43 (Suppl. 1), 27–36. 10.1111/apt.13436. [DOI] [PubMed] [Google Scholar]
- Podszun M. C.; Jakobi M.; Birringer M.; Weiss J.; Frank J. The long chain alpha-tocopherol metabolite alpha-13′-COOH and gamma-tocotrienol induce P-glycoprotein expression and activity by activation of the pregnane X receptor in the intestinal cell line LS 180. Mol. Nutr. Food Res. 2017, 61, 1600605. 10.1002/mnfr.201600605. [DOI] [PubMed] [Google Scholar]
- Galli F.; Polidori M. C.; Stahl W.; Mecocci P.; Kelly F. J. Vitamin E biotransformation in humans. Vitam. Horm. 2007, 76, 263–280. 10.1016/S0083-6729(07)76009-0. [DOI] [PubMed] [Google Scholar]
- Schubert M.; Kluge S.; Schmölz L.; Wallert M.; Galli F.; Birringer M.; Lorkowski S. Long-chain metabolites of vitamin E: metabolic activation as a general concept for lipid-soluble vitamins?. Antioxidants 2018, 7, E10. 10.3390/antiox7010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birringer M.; Drogan D.; Brigelius-Flohe R. Tocopherols are metabolized in HepG2 cells by side chain omega-oxidation and consecutive beta-oxidation. Free Radical Biol. Med. 2001, 31, 226–232. 10.1016/S0891-5849(01)00574-3. [DOI] [PubMed] [Google Scholar]
- Birringer M.; Pfluger P.; Kluth D.; Landes N.; Brigelius-Flohe R. Identities and differences in the metabolism of tocotrienols and tocopherols in HepG2 cells. J. Nutr. 2002, 132, 3113–3118. 10.1093/jn/131.10.3113. [DOI] [PubMed] [Google Scholar]
- Marinelli R.; Torquato P.; Bartolini D.; Mas-Bargues C.; Bellezza G.; Gioiello A.; Borras C.; Fallarino F.; Mani S.; Sidoni A.; Viña J.; Galli F., manuscript in preparation.
- Mazzini F.; Betti M.; Netscher T.; Galli F.; Salvadori P. Configuration of the vitamin E analogue garcinoic acid extracted from Garcinia Kola seeds. Chirality 2009, 21, 519–524. 10.1002/chir.20630. [DOI] [PubMed] [Google Scholar]
- Wallert M.; Bauer J.; Kluge S.; Schmölz L.; Chen Y. C.; Ziegler M.; Searle A. K.; Maxones A.; Schubert M.; Thürmer M.; Pein H.; Koeberle A.; Werz O.; Birringer M.; Peter K.; Lorkowski S. The vitamin E derivative garcinoic acid from Garcinia kola nut seeds attenuates the inflammatory response. Redox Biol. 2019, 24, 101166. 10.1016/j.redox.2019.101166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birringer M.; Lington D.; Vertuani S.; Manfredini S.; Scharlau D.; Glei M.; Ristow M. Proapoptotic effects of long-chain vitamin E metabolites in HepG2 cells are mediated by oxidative stress. Free Radical Biol. Med. 2010, 49, 1315–1322. 10.1016/j.freeradbiomed.2010.07.024. [DOI] [PubMed] [Google Scholar]
- Terashima K.; Takaya Y.; Niwa M. Powerful antioxidative agents based on garcinoic acid from Garcinia kola. Bioorg. Med. Chem. 2002, 10, 1619–1625. 10.1016/S0968-0896(01)00428-X. [DOI] [PubMed] [Google Scholar]
- Mostarda S.; Filipponi P.; Sardella R.; Venturoni F.; Natalini B.; Pellicciari R.; Gioiello A. Glucuronidation of bile acids under flow conditions: design of experiments and Koenigs–Knorr reaction optimization. Org. Biomol. Chem. 2014, 12, 9592–9600. 10.1039/C4OB01911C. [DOI] [PubMed] [Google Scholar]
- Xue Y.; Chao E.; Zuercher W. J.; Willson T. M.; Collins J. L.; Redinbo M. R. Crystal structure of the PXR-T1317 complex provides a scaffold to examine the potential for receptor antagonism. Bioorg. Med. Chem. 2007, 15, 2156–2166. 10.1016/j.bmc.2006.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou W.; Mukherjee S.; Li H.; Venkatesh M.; Wang H.; Kortagere S.; Peleg A.; Chilimuri S. S.; Wang Z. T.; Feng Y.; Fearon E. R.; Mani S. Alleviation of gut inflammation by Cdx2/Pxr pathway in a mouse model of chemical colitis. PLoS One 2012, 7, e36075 10.1371/journal.pone.0036075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez-Campoy A.; Todd M. J.; Freire E. HIV-1 protease inhibitors: enthalpic versus entropic optimization of the binding affinity. Biochemistry 2000, 39, 2201–2207. 10.1021/bi992399d. [DOI] [PubMed] [Google Scholar]
- Ruben A. J.; Kiso Y.; Freire E. Overcoming roadblocks in lead optimization: a thermodynamic perspective. Chem. Biol. Drug Des. 2006, 67, 2–4. 10.1111/j.1747-0285.2005.00314.x. [DOI] [PubMed] [Google Scholar]
- Zhou C.; Poulton E. J.; Grün F.; Bammler T. K.; Blumberg B.; Thummel K. E.; Eaton D. L. The dietary isothiocyanate sulforaphane is an antagonist of the human steroid and xenobiotic nuclear receptor. Mol. Pharmacol. 2007, 71, 220–229. 10.1124/mol.106.029264. [DOI] [PubMed] [Google Scholar]
- Leist M.; Raab B.; Maurer S.; Rösick U.; Brigelius-Flohé R. Conventional cell culture media do not adequately supply cells with antioxidants and thus facilitate peroxide-induced genotoxicity. Free Radical Biol. Med. 1996, 21, 297–306. 10.1016/0891-5849(96)00045-7. [DOI] [PubMed] [Google Scholar]
- Goodwin B.; Hodgson E.; Liddle C. The orphan human pregnane X receptor mediates the transcriptional activation of CYP3A4 by rifampicin through a distal enhancer module. Mol. Pharmacol. 1999, 56, 1329–1339. 10.1124/mol.56.6.1329. [DOI] [PubMed] [Google Scholar]
- Toriyabe T.; Nagata K.; Takada T.; Aratsu Y.; Matsubara T.; Yoshinari K.; Yamazoe Y. Unveiling a new essential cis element for the transactivation of the CYP3A4 gene by xenobiotics. Mol. Pharmacol. 2009, 75, 677–684. 10.1124/mol.108.050575. [DOI] [PubMed] [Google Scholar]
- Torquato P.; Bartolini D.; Giusepponi D.; Saluti G.; Russo A.; Barola C.; Birringer M.; Galarini R.; Galli F. a-13′-OH is the main product of a-tocopherol metabolism and influences CYP4F2 and PPAR- Gene expression in HepG2 human hepatocarcinoma cells. Free Radical Biol. Med. 2016, 96, S19–S20. 10.1016/j.freeradbiomed.2016.04.159. [DOI] [Google Scholar]
- Russo A.; Bartolini D.; Torquato P.; Giusepponi D.; Barola C.; Galarini R.; Birringer M.; Lorkowski S.; Galli F. CYP4F2 repression and a modified alpha-tocopherol (vitamin E) metabolism are two independent consequences of ethanol toxicity in human hepatocytes. Toxicol. In Vitro 2017, 40, 124–133. 10.1016/j.tiv.2016.12.014. [DOI] [PubMed] [Google Scholar]
- Landes N.; Pfluger P.; Kluth D.; Birringer M.; Rühl R.; Böl G. F.; Glatt H.; Brigelius-Flohé R. Vitamin E activates gene expression via the pregnane X receptor. Biochem. Pharmacol. 2003, 65, 269–273. 10.1016/S0006-2952(02)01520-4. [DOI] [PubMed] [Google Scholar]
- Pein H.; Ville A.; Pace S.; Temml V.; Garscha U.; Raasch M.; Alsabil K.; Viault G.; Dinh C.-P.; Guilet D.; Troisi F.; Neukirch K.; König S.; Bilancia R.; Waltenberger B.; Stuppner H.; Wallert M.; Lorkowski S.; Weinigel C.; Rummler S.; Birringer M.; Roviezzo F.; Sautebin L.; Helesbeux J.-J.; Séraphin D.; Mosig A. S.; Schuster D.; Rossi A.; Richomme P.; Werz P. O.; Koeberle A. Endogenous metabolites of vitamin E limit inflammation by targeting 5-lipoxygenase. Nat. Commun. 2018, 9, 3834. 10.1038/s41467-018-06158-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torquato P.; Giusepponi D.; Alisi A.; Galarini R.; Bartolini D.; Piroddi M.; Goracci L.; Di Veroli A.; Cruciani G.; Crudele A.; Nobili V.; Galli F. Nutritional and lipidomics biomarkers of docosahexaenoic acid-based multivitamin therapy in pediatric NASH. Sci. Rep. 2019, 9, 2045. 10.1038/s41598-018-37209-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torquato P.; Bartolini D.; Giusepponi D.; Piroddi M.; Sebastiani B.; Saluti G.; Galarini R.; Galli F. Increased plasma levels of the lipoperoxyl radical-derived vitamin E metabolite α-tocopheryl quinone are an early indicator of lipotoxicity in fatty liver subjects. Free Radical Biol. Med. 2019, 131, 115–125. 10.1016/j.freeradbiomed.2018.11.036. [DOI] [PubMed] [Google Scholar]
- Terashima K.; Shimamura T.; Tanabayashi M.; Aqil M.; Akinniyi J. A.; Niwa M. Constituents of the seeds of Garcinia Kola: two new antioxidants, garcinoic acid and garcinal. Heterocycles 1997, 45, 1559–1566. 10.3987/COM-97-7854. [DOI] [Google Scholar]
- Wang W.; Prosise W. W.; Chen J.; Taremi S. S.; Le H. V.; Madison V.; Cui X.; Thomas A.; Cheng K. C.; Lesburg C. A. Construction and characterization of a fully active PXR/SRC-1 tethered protein with increased stability. Protein Eng., Des. Sel. 2008, 21, 425–433. 10.1093/protein/gzn017. [DOI] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010, 66, 125–132. 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn M. D.; Ballard C. C.; Cowtan K. D.; Dodson E. J.; Emsley P.; Evans P. R.; Keegan R. M.; Krissinel E. B.; Leslie A. G. W.; McCoy A.; McNicholas S. J.; Murshudov G. N.; Pannu N. S.; Potterton E. A.; Powell H. R.; Read R. J.; Vagin A.; Wilson K. S. Overview of the CCP4 suite and current developments. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2011, 67, 235–242. 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo G.; Passeri D.; De Franco F.; Ciaccioli G.; Donadio L.; Rizzo G.; Orlandi S.; Sadeghpour B.; Wang X. X.; Jiang T.; Levi M.; Pruzanski M.; Adorini L. Functional characterization of the semisynthetic bile acid derivative INT-767, a dual farnesoid X receptor and TGR5 agonist. Mol. Pharmacol. 2010, 78, 617–630. 10.1124/mol.110.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace B. D.; Betts L.; Talmage G.; Pollet R. M.; Holman N. S.; Redinbo M. R. Structural and functional analysis of the human nuclear xenobiotic receptor PXR in complex with RXRα. J. Mol. Biol. 2013, 425, 2561–2577. 10.1016/j.jmb.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- Bartolini D.; Commodi J.; Piroddi M.; Incipini L.; Sancineto L.; Santi C.; Galli F. Glutathione S-transferase pi expression regulates the Nrf2-dependent response to hormetic diselenides. Free Radical Biol. Med. 2015, 88, 466–480. 10.1016/j.freeradbiomed.2015.06.039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.