Table III.
Data collection and refinement statistics for scTGPp1-1
| Data collection statistics | |
| Wavelength (Å) | 1.0 |
| Resolution range (Å) | 50–2.50 (2.50–2.54) |
| Space group | P41212 |
| Unit-cell parameters (Å) | a = b = 103.96, c = 122.74 |
| Total reflections | 3 33 231 |
| Unique reflections | 24 030 (1177) |
| Completeness (%) | 100.0 (100.0) |
| Multiplicity | 13.9 (13.7) |
| Rmergea (%) | 5.9 (100.0) |
| Rp.i.mb | 0.019 (0.467) |
| Rmeas/Rr.i.mb | 0.062 (1.000) |
| Wilson B factor | 34.51 |
Average I/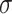 (I) (I) |
40.3 (1.7) |
| Refinement statistics | |
| Resolution (Å) | 50.00–2.50 (2.59–2.50) |
| Completeness (%) | 91.85 (75.20) |
| Working reflections | 21 973 (1771) |
| Test reflections | 1839 (150) |
| Rworkc (%) | 0.1630 (0.2343) |
| Rfreec (%) | 0.2111 (0.2711) |
| No. of waters | 136 |
| No. of other solvent molecules | 2 GOL |
| R.M.S. deviations from ideal geometry | |
| Bonds (Å) | 0.006 |
| Angles (°) | 0.84 |
| Average B factors (Å2) | 43.74 |
| Ramachandran analysis by MOLPROBITY | |
| Residues in favored regions (%) | 98.90 |
| Residues in allowed regions (%) | 1.10 |
Values in parentheses represent the outermost resolution shell.
a R merge = ΣhklΣi|Ii(hkl) − <I(hkl)>|/ΣhklΣiIi(hkl) where Ii(hkl) is the intensity of the ith observation and <I(hkl)> is the mean intensity of the reflections. The values are for unmerged Friedel pairs.
b R p.i.m. (precision-indicating Rmerge) and Rmeas/Rr.i.m. (redundancy-independent Rmerge).
cThe crystallographic R factor R = Σhkl| |Fobs| − |Fcal| |/Σhkl |Fobs|; Rfree = Σhkl | |Fobs| − |Fcal| |/Σhkl |Fobs| where all reflections belong to a test set of randomly selecteddata.
