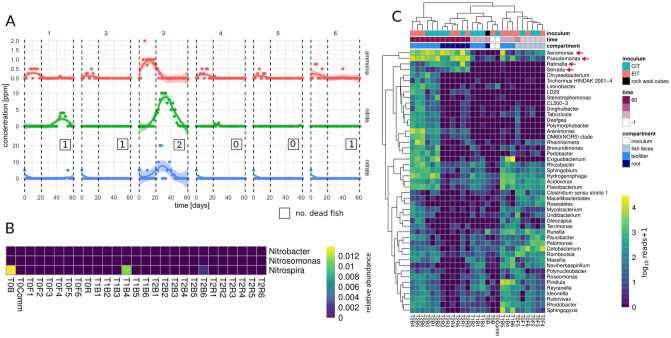
Fig 2. Nitrogen cycling and microbial composition.
(A) Shown are measurements for ammonia, nitrite and nitrate throughout the experiment. Colored lines denote smooth fit from a LOESS regression and filled areas denote 95% confidence intervals of the regression. Numbers of the panels denote tanks. Tanks 1–3 are CIT whereas tanks 4–6 are EIT. Dashed lines denote the three time points used for microbiome sampling. Boxed numbers denote the number of fish that died in the tank. (B) Relative abundance (fraction of total reads per sample) of known nitrifying taxa. (C) Abundances of ubiquitous bacterial genera across the samples (present in at least 2 samples at an abundance >300 reads). Colors of cells denote the normalized abundance on a base 10 log-scale. Sample names are composed of sampling group ID (e.g. T1), compartment (B = biofilter, F = fish feces, R = root, Comm = commercial inoculum) and tank number (1–6). Orange arrows denote genera of interest. Black fill color denotes initial root sample from rock wool cubes (not part of inoculation strategy).