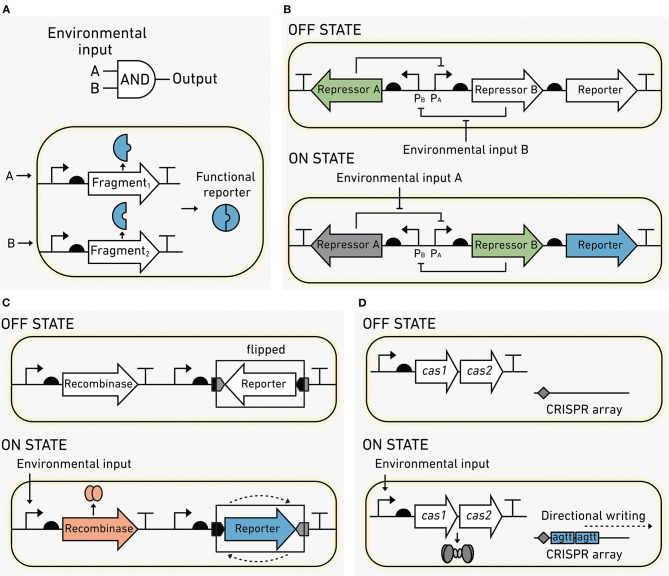
Figure 8.
Processing modules can increase biosensor sophistication. (A) Two-input AND gates can be created by using modules that encode protein in fragments. The output only occurs when the first AND second fragment are produced. (B) Flexible DNA memory using a toggle switch. In the OFF state, transcriptional repressor A blocks the production of the repressor B and the reporter output. Upon sensing environmental input A, the circuits flip to an ON state where repressor A is momentarily dissociated from promoter PA, allowing the production of repressor B and the output. Repressor B blocks production of repressor A, stabilizing this ON state. The switch can be flipped back to the OFF state upon sensing of environmental input B. (C) Fixed DNA memory built using recombinases. In the OFF state, no recombinase is made, and the reporter DNA sequence is antiparallel to the regulatory elements required for expression. In the ON state, an environmental input induces recombinase production, which binds a pair of DNA sequences flanking the reporter and inverts the DNA such that the reporter is parallel to the regulatory elements. This flipping of the DNA leads to output production. (D) Fixed DNA memory that uses CRISPR. Input sensing is coupled to the expression of Cas1/2, which incorporates short DNA spacers into a CRISPR array at a rate that is proportional to the input exposure.