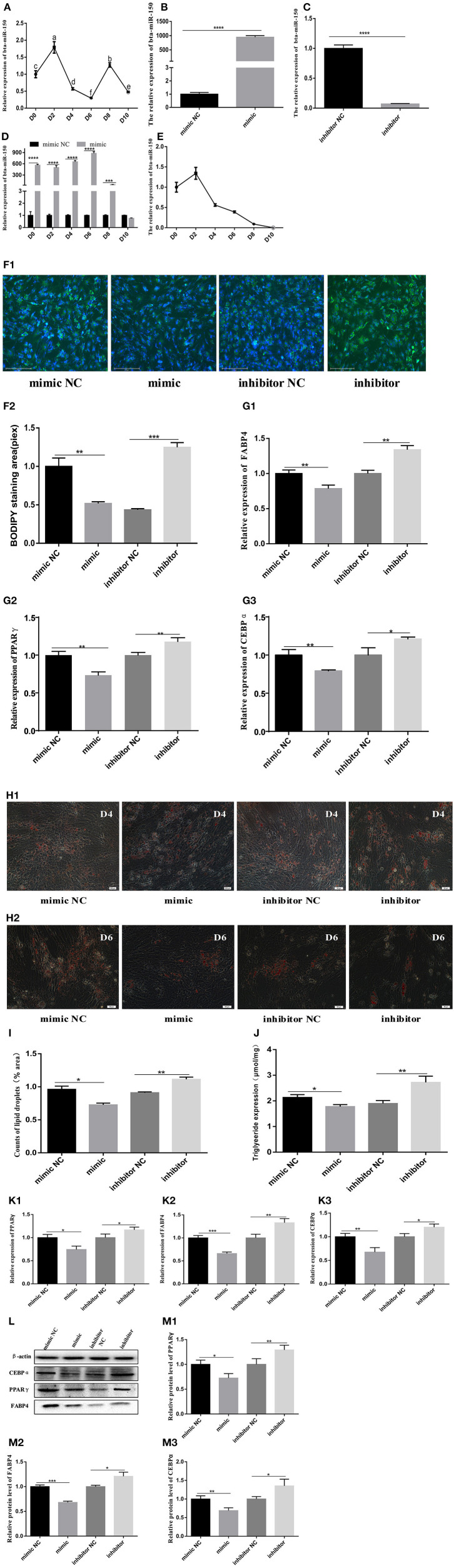
Figure 3.
Differentiation of pre-adipocytes from Qinchuan cattle. (A) Background expression of bta-miR-150 on different days after inducting differentiation in Qinchuan cattle adipocytes. (B) bta-miR-150 mimic transfection efficiency. (C) bta-miR-150 inhibitor transfection efficiency. (D) Overexpression of bta-miR-150 treated with miR-150 mimic in 10 days. (E) Changes of bta-miR-150 content in Qinchuan cattle adipocytes treated with miR-150 mimics. (F1,F2) After transfection of miR-150 mimic, miR-150 inhibitor, or negative control, the adipocytes were stained with Bodipy and quantified on the second day of differentiation; photos were taken with an Evos-fl-auto2 automatic living cell fluorescence microscopy imaging system (Thermo Fisher, USA). (G1–G3) Changes in mRNA levels of adipose differentiation marker genes on the second day of differentiation (FABP4, PPARγ, and CEBPα). (H1) Oil red O staining of mimics, inhibitors, and their respective internal controls on day 4; the photos were taken with an Olympus Image IX71 microscope (Olympus, Japan). (H2) bta-miR-150 mimic treatment on day 6, Oil red O staining of substances, inhibitors, and their respective internal controls; the photos were taken with an Olympus Image IX71 microscope (Olympus, Japan). (I) Statistics of lipid droplet content on day 4 of differentiation, using Image J software. (J) Triglyceride content on day 4 of differentiation. (K1–K3) Changes in mRNA levels of fat differentiation marker genes (FABP4, PPARγ, CEBPα) on the 4th day. (L) Protein map of fat differentiation marker genes (FABP4, PPARγ, CEBPα). (M1–M3) Quantitative processing of fat differentiation marker gene protein results (FABP4, PPARγ, CEBPα); protein levels were quantified and analyzed by Image J software. Error bars represent mean ± SD. n = 3 replicates. *denotes significance according to Student's t-test, *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.