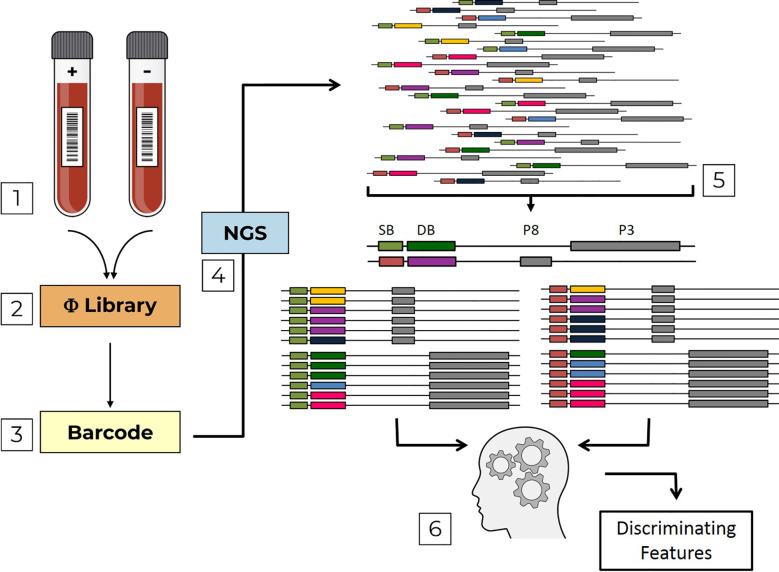
Figure 1.
Pipeline flow chart. The pipeline is composed of sequential steps; (1) Serum samples, which are either positive (+) or negative (-) with respect to a specific infection are used to screen Domain-Scan phage-displayed libraries (2) During PCR sample preparation, sample barcodes are incorporated into the sequence affinity selected phages (3) that are then sent to NGS (4). Erroneous DNA reads are filtered out. Then, the remaining reads are parsed (5) first by sample barcodes (SB, green and orange) and then by domain barcodes (DB, yellow, violet, blue, dark-green, light blue and pink). The two libraries (PVIII and PIII) are analyzed by a machine-learning algorithm (6) to identify discriminating features.