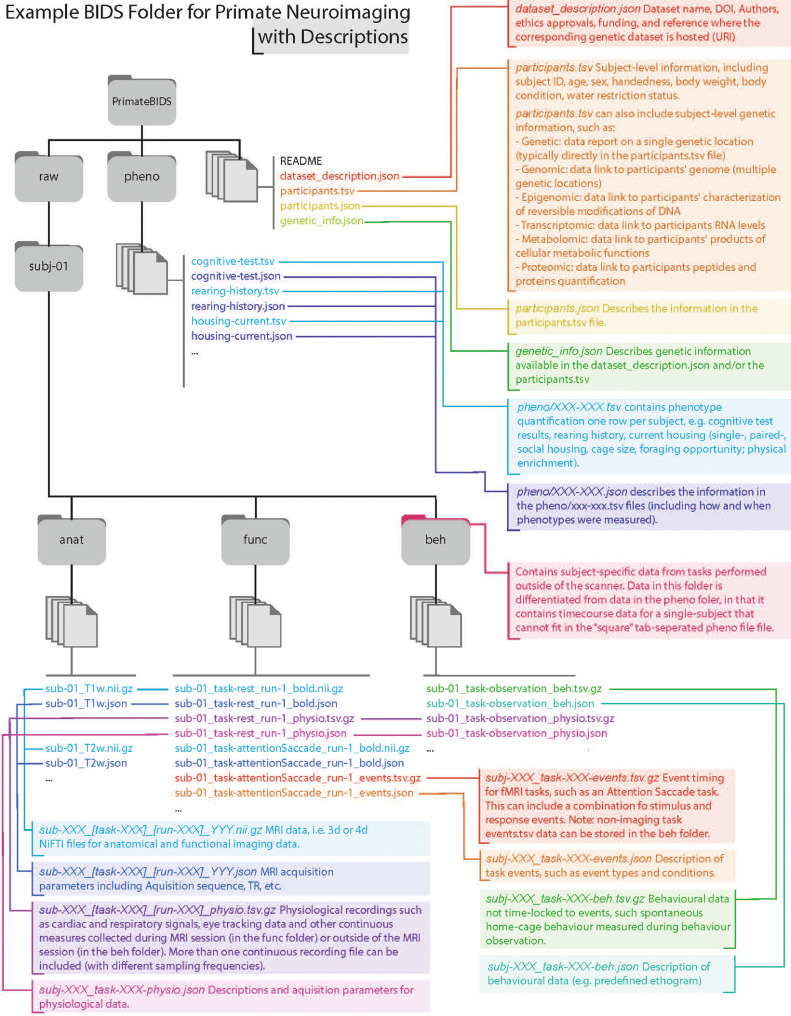
Fig.2.
Example BIDS Folder for Primate Neuroimaging with Metadata. The BIDS format describes a unified structure for sharing brain imaging data in files and folders (grey). Here we show the organization of an example BIDS dataset, with file descriptions (connected to files with coloured lines). Briefly, each study has a top-level directory (e.g. PrimateBIDS), which contains folders for raw data (raw) and phenotype information (pheno), along with descriptor files where researchers can share descriptions of the subjects, dataset, and accompanying genetic information. The pheno folder houses across-subject data (.tsv) and descriptions of those data (.json). The raw folder contains a folder for each subject, e.g. subj-XX. Each subject-folder, in turn, contains anat, func, and beh folders, which contain anatomical, functional, and behavioural data, respectively. Similar to the pheno folder, each of these folders contains data (e.g. .nii.gz or .tsv) with an accompanying descriptor file (.json), which describes relevant acquisition/quantification parameters. Individual subject folders can be used to share any data that cannot be easily summarized into an across-subject tabular .tsv file in the pheno folder.