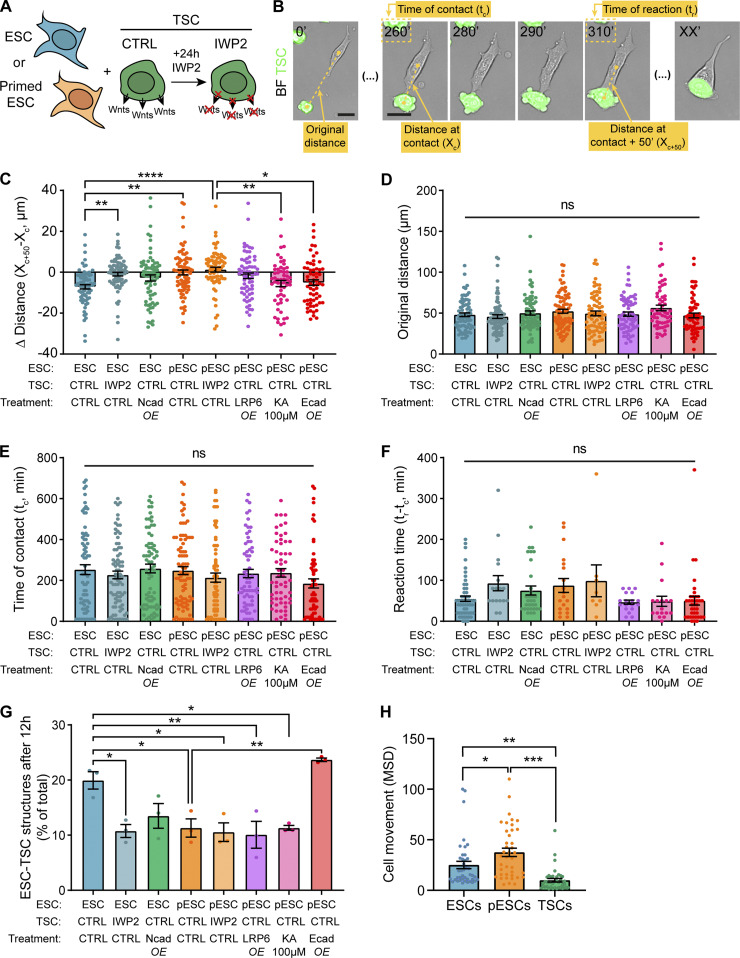
Figure S2.
Quantification of ESC or pESC interaction with TSCs. (A) Schematic depicting the experimental conditions. (B) Representative annotated frames of a time-lapse live-cell imaging showing an ESC contacting a TSC with a cytoneme and pairing with it. TSCs express GFP and are labeled in green. Annotations refer to measurements in C–F, as follows: “Original distance” between the cells at t = 0’; distance between the cells at the time of cytoneme-mediated contact (“Distance at contact”; Xc); time at initial cytoneme-mediated contact (“Time at contact”; tc); distance between cells 50 min after contact (“Distance at contact + 50 minutes”; Xc+50; found empirically to be enough to capture the behavior of the cells after initial contact); time at which cell–cell pairing is established (“Time of reaction”; tr; only quantified for reactive interactions). Time is in minutes. Scale bars, 20 µm. BF, brightfield. (C) Quantification of the differential between the distance at contact (Xc) and distance at contact + 50 min (Xc+50) expressed in micrometers, for all conditions. n ≥ 58 cells pooled from ≥3 independent experiments. (D) Quantification of the original ESC–TSC distance (in micrometers) for all conditions. n ≥ 58 cells pooled from ≥3 independent experiments. (E) Quantification of the time of ESC–TSC contact through a cytoneme (in minutes) for all conditions. n ≥ 58 cells pooled from ≥3 independent experiments. (F) Quantification of reaction time between ESCs and TSCs, calculated as the differential between time at contact (tc) and time at reaction (tr) in minutes, for all conditions. Reaction time is only calculated for cells that react (according to Fig. 3 A). n ≥ 8 cells from ≥3 independent experiments. (G) Quantification of the number of mixed ESC–TSC structures after 12 h co-culture, presented as percentage of total TSC clusters. n = 3, ≥66 total structures per n. (H) Quantification of cell movement for ESCs (blue), pESCs (orange), and TSCs (green), presented as mean squared displacement (MSD). n = 40 cells pooled from ≥3 experiments. For C–H, bars indicate mean, and error bars are SEM. Symbols indicate statistical significance calculated by one-way ANOVA with Tukey’s multiple comparison tests: ns, nonsignificant, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. CTRL, control; Ecad, E-cadherin; KA, kainate; Ncad, N-cadherin; OE, overexpression.