DNA methylation plays a critical role in innate immune response, but the underlying mechanisms have not been clarified. The authors found a single-nucleotide methylation controls IFN-I induction and antiviral immunity via regulating IRF3 recruitment both in human and mouse.
Abstract
Frequent outbreaks of viruses have caused a serious threat to public health. Previous evidence has revealed that DNA methylation is correlated with viral infections, but its role in innate immunity remains poorly investigated. Additionally, DNA methylation inhibitors promote IFN-I by upregulating endogenous retrovirus; however, studies of intrinsically demethylated tumors do not support this conclusion. This study found that Uhrf1 deficiency in myeloid cells significantly upregulated Ifnb expression, increasing resistance to viral infection. We performed whole-genome bisulfite sequencing and found that a single-nucleotide methylation site in the Ifnb promoter region disrupted IRF3 recruitment. We used site-specific mutant knock-in mice and a region-specific demethylation tool to confirm that this methylated site plays a critical role in regulating Ifnb expression and antiviral responses. These findings provide essential insight into DNA methylation in the regulation of the innate antiviral immune response.
Graphical Abstract
Introduction
With the outbreak of H1N1, polio, Ebola, Zika, and COVID-19 in recent years, infections caused by viruses have received increased attention (Dong et al., 2020). Additionally, seasonal influenza causes serious morbidity and mortality worldwide and huge economic losses (Bommarito and Fry, 2019). Therefore, it is important to clarify the detailed mechanism of antiviral infection. The host produces type I IFN (IFN-I) and type II IFN (IFN-II) to defend against viral infection and regulate the immune response. The production of IFN-I involves the recognition of pathogen-associated molecular patterns by host pattern recognition receptors. Viral RNA is recognized by TLRs and RIG-I–like receptors. Additionally, DNA sensors, including cyclic GMP-AMP, AIM2, and IFI16, recognize viral DNA (Paludan and Bowie, 2013). After recognizing viral RNA and DNA, these sensors trigger the activation of the transcription factors (TFs) NF-κB, IRF3, and IRF7 via distinct adaptor proteins, such as TRIF, mitochondrial antiviral signaling, and STING, leading to the induction of IFN-I (Takeuchi and Akira, 2010). Once produced, IFN-I results in the phosphorylation of STAT1 and STAT2, leading to the expression of IFN-stimulated genes (ISGs; Lee and Ashkar, 2018; Wang et al., 2017). IFN-II is mainly produced by NK cells during antiviral infection and is dependent on STAT4 phosphorylation. Subsequently, IFN-II leads to the homodimerization and phosphorylation of STAT1 (Lee and Ashkar, 2018).
Although signal transduction and TFs described above have been shown to play important roles in the production of IFN in recent years, an increasing number of studies have found that epigenetics also regulates the production of IFN (Selinger and Reiniš, 2018; Shestakova, 2015; Aune et al., 2013). DNA methylation is one of the most important and first-discovered epigenetic modifications (Hotchkiss, 1948) and plays a pivotal role in X chromosome inactivation, genomic imprinting, and transposable element repression (Bommarito and Fry, 2019). DNA methylation is established by DNMT3a and DNMT3b during embryogenesis (Okano et al., 1999). Next, ubiquitin-like with plant homeodomain and RING finger domain 1 (UHRF1; also known as NP95 in mice and ICBP90 in humans) recruits DNMT1 to hemimethylated DNA to maintain DNA methylation during mitosis (Bostick et al., 2007). DNA demethylation is performed by ten-eleven translocation (TET) methylcytosine dioxygenase family members (TET1, TET2, and TET3), followed by thymine DNA glycosylase excision and the base excision repair (He et al., 2011).
DNA demethylation affects various physiological functions and is involved in diverse biological processes, including mammalian development and the stress response in normal individuals. Tumor cells have been suggested to contain aberrant DNA methylation with global hypomethylation and local hypermethylation at the promoters of tumor suppressor genes (Baylin and Jones, 2016). DNA methyltransferase inhibitors have been applied in myelodysplastic syndrome and acute myeloid leukemia, but not all patients benefit from this inhibitor therapy (Issa and Kantarjian, 2009; Jones et al., 2019). DNA methylation also shapes immune cell development, differentiation, and physiological function in host defense (Morales-Nebreda et al., 2019; Suarez-Alvarez et al., 2012). T helper cell (Th cell) differentiation is regulated by DNA demethylation at the relevant gene loci, which are often reversely methylated in other lineages (Ichiyama et al., 2015). Infection with Mycobacterium tuberculosis in dendritic cells was shown to induce rapid DNA demethylation, leading to the activation of most immune TFs (Pacis et al., 2015). DNA methyltransferases and demethylases have been reported to be involved in the immune response. DNMT1 is required for the fate and function of T cells, as demonstrated by the defective development of early double-negative or double-positive DNMT1-deficient thymocytes (Lee et al., 2001). DNMT1 is also required for the fate of regulatory T cells (T reg cells) to prevent the development of lethal autoimmunity (Wang et al., 2013). DNMT3a not only regulates the plasticity of CD4+ T cell cytokine expression via the methylated Ifng promoter and Il13 promoter (Gamper et al., 2009; Thomas et al., 2012; Yu et al., 2012), but also restricts the development of memory precursor cells and allows normal terminal CD8+ T effector cell differentiation (Ladle et al., 2016). TET2 and TET3 regulate lineage differentiation by promoting T-bet and ThPOK expression by removing methylation modifications at these gene loci in T cells (Tsagaratou et al., 2017). TET2 has also been reported to limit CD8+ T cell memory differentiation (Carty et al., 2018). In T reg cells, TET1 and TET2 catalyze Foxp3 demethylation to stabilize Foxp3 expression and maintain immune homeostasis (Yang et al., 2015). Additionally, TET2 and TET3 are involved in T reg cell maintenance (Nakatsukasa et al., 2019; Yue et al., 2019).
DNA methylation is essential for innate immunity, particularly for the induction of multiple cytokines. DNMT1 and DNMT3b promote M1 macrophage activation by promoting the methylation of the KLF4 and PPAR-γ promoters (Davis and Gallagher, 2019; Tang et al., 2019; Wang et al., 2016; Yang et al., 2014; Yu et al., 2016). TET2-deficient macrophages show an increase in LPS-induced and spontaneous inflammation (Cull et al., 2015, 2017). These regulators have also been reported to regulate cytokine production. TET1 promotes TNF-α expression in macrophages, possibly by regulating 5hmC in the TNF-α promoter (Sun et al., 2019). TET2 recruits histone deacetylase 2 (HDAC2) to specifically repress IL-6 independent of DNA methylation in innate myeloid cells (Zhang et al., 2015). Another study also revealed that TET2 loss increased Il1b and Il6 expression in macrophages (Cull et al., 2017). DNMT1 and UHRF1 mutants in zebrafish embryos induced an IFN-I response by demethylating and stabilizing class I retrotransposons, and the promoters of ifnphi1 and ifnphi4 were completely demethylated in both WT and mutant embryos (Chernyavskaya et al., 2017). However, current evidence suggests that TET3 decreases IFN-I transcription by recruiting HDAC1 and the transcriptional corepressor SIN3A to the Ifnb1 promoter, events that occur independent of DNA demethylation (Zhang et al., 2015).
Although this evidence suggests that DNA methylation has an important regulatory effect on the innate immune response, the specificity and underlying molecular mechanisms of DNA methylation have not yet been clarified; this lack of knowledge hinders the development of therapeutic methods targeting DNA methylation to treat infectious diseases and tumors. In the present study, we identified an unexpected role of Uhrf1 in the pathogenic characteristics of different influenza (flu) virus strains. Compared with H5N1, the H1N1 influenza virus causes a significant reduction in Uhrf1 mRNA levels and increases IFN-I production. Genetic deficiency further revealed a specific negative role of Uhrf1 in virus-induced death and IFN-I induction without affecting the activities of critical signaling components in this pathway. Whole-genome bisulfite sequencing revealed that Uhrf1-regulated production of IFN-I is mainly mediated by single and specific methylation of the Ifnb promoter. Using dCas9-Tet1, we demonstrated that targeted demethylation at a single methylation site enhances IRF3 recruitment and promotes IFN-I expression. These findings reveal a surprising mechanism that controls IFN-I induction and establish a crucial therapeutic tool by targeting DNA methylation in antiviral innate immunity.
Results
Influenza virus infection causes a reduction in Uhrf1-mediated DNA methylation
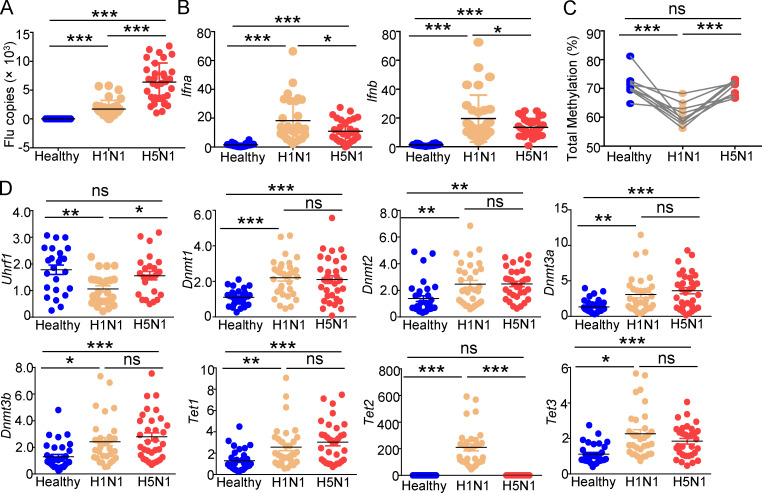
Because distinct strains of influenza virus infections cause different clinical pathogenic characteristics, we collected peripheral blood mononuclear cells (PBMCs) from 32 healthy donors infected with either the H1N1 or H5N1 virus. Compared with H1N1 infection, H5N1 infection led to a significant increase in viral loads and a reduction in IFN-I induction (Fig. 1, A and B). Epigenetic modifications play important roles in gene expression, in which DNA methylation has been proven to control gene transcription by affecting chromatin assembly and specific TF recruitment. Therefore, we further compared the levels of DNA methylation in the whole genome of PBMCs after infection with H1N1 or H5N1. Consistent with the increased IFN-I induction induced by H1N1 infection, DNA methylation was also significantly reduced in H1N1-infected PBMCs compared with that in H5N1-infected PBMCs (Fig. 1 C).
Figure 1.
Distinct strains of influenza virus lead to different levels of DNA methylation and IFN-I induction. (A and B) Viral loads (A) and mRNA levels of Ifnb (B) induced by different influenza viruses (1 MOI for 6 h) in PBMCs from healthy donors (n = 32), as measured by qPCR. (C) Pyrosequencing results of the methylation levels in the whole genome of PBMCs from healthy donors infected with different influenza virus strains. (D) The expression of the indicated genes in PBMCs from healthy donors (n = 32) infected with different influenza viruses (1 MOI for 6 h) in vitro was measured by qPCR. The data from the qPCR assay are presented as fold changes relative to the actin mRNA levels. All data are representative of at least three independent experiments. The data are presented as means ± SEMs. The significance of differences was determined using a t test. *, P < 0.05; **, P < 0.01; ***, P < 0.005.
DNMTs are the writers and maintainers of DNA methylation. By contrast, TETs erase this methylation (Lyko, 2018; Rasmussen and Helin, 2016). Thus, we evaluated the expression of these critical molecules involved in DNA methylation, such as Uhrf1, Dnmt1, Dnmt3, and Tet1-3. The results demonstrated that although influenza virus infection led to the upregulation of TET expression, no or opposite differences were observed between the H1N1- or H5N1-infected groups, except for Tet2 (Fig. 1 D). Additionally, these infections enhanced the expression of Dnmt1, Dnmt2, and Dnmt3a-b, but not that of Uhrf1. Consistent with the higher induction of Ifnb, H1N1 infection inhibited the expression of Uhrf1 more significantly than H5N1 infection (Fig. 1 D). These data imply a functional role of Uhrf1-mediated DNA methylation in the induction of IFN-I by different influenza strains and antiviral immune responses.
Uhrf1 plays a negative role in antiviral innate immunity
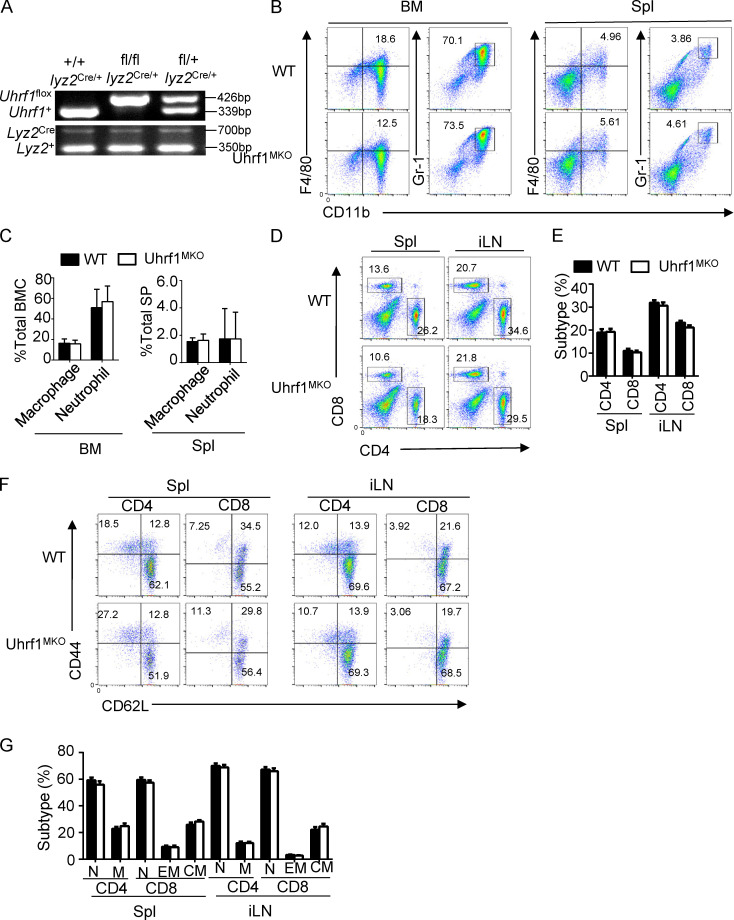
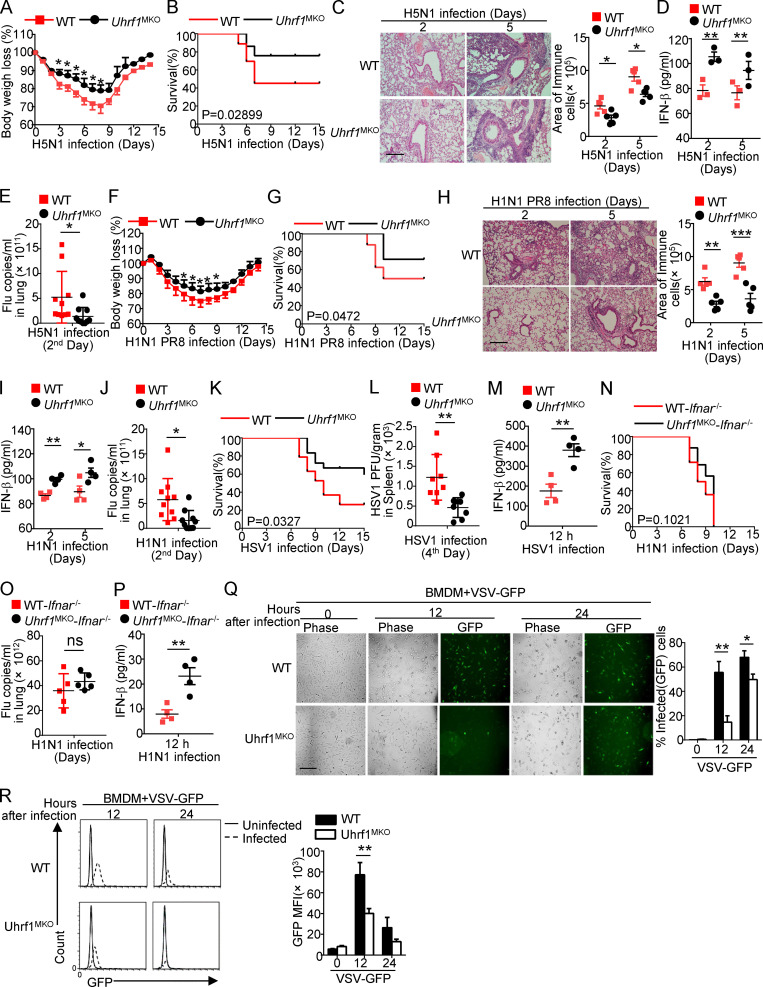
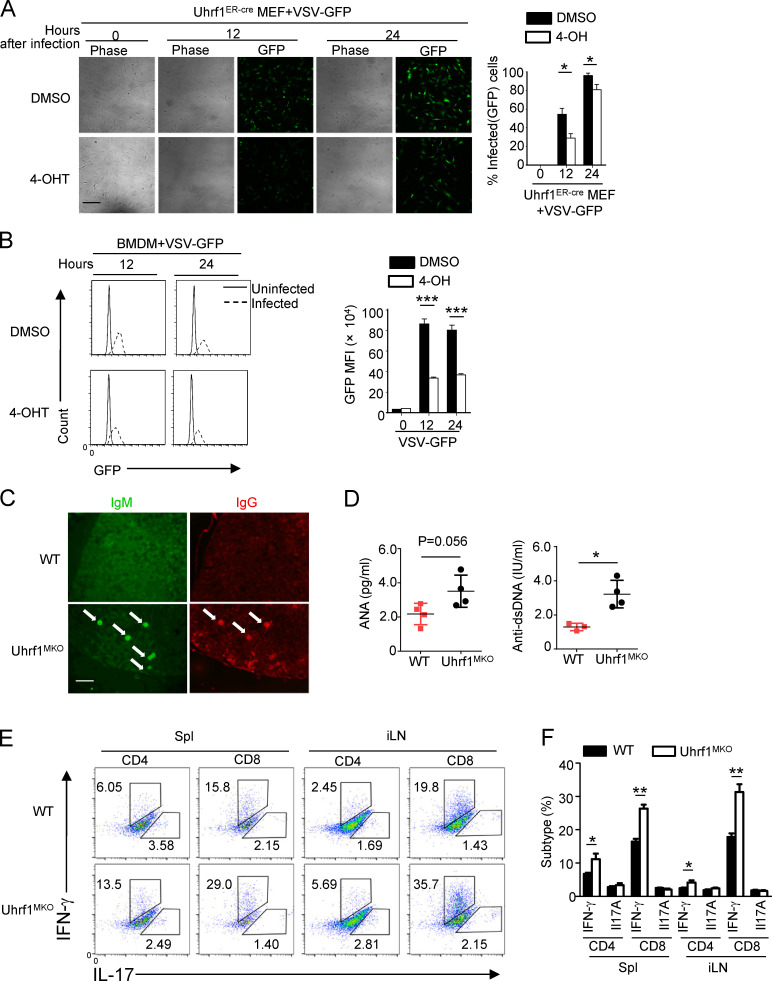
To characterize the role of Uhrf1 in influenza A virus (IAV) infection in vivo, we first crossed Uhrf1flox mice with Lyz2-Cre mice to produce myeloid cell–specific Uhrf1-KO (Uhrf1MKO) mice (Fig. S1 A). Compared with WT mice, the Uhrf1MKO mice displayed no differences in the frequencies of myeloid cells in bone marrow (BM) and spleen (Fig. S1, B and C). The proportion of splenic T lymphocytes in Uhrf1MKO mice was comparable to that in WT mice (Fig. S1, D and E). Uhrf1 deficiency in myeloid cells had no effect on splenic T cell homeostasis (Fig. S1, F and G). We then infected WT and Uhrf1MKO mice i.n. using the H5N1 strain at a 50% tissue culture infectious dose (TCID50) of 10 and monitored the body weight loss and survival ratio for 2 wk. Compared with their WT littermates, Uhrf1MKO mice exhibited less body weight loss (Fig. 2 A) and a significant reduction in the IAV-induced mouse mortality rate (Fig. 2 B). As expected, Uhrf1MKO mice exhibited significantly alleviated immune cell infiltration and injury in the lungs, including decreased alveolar wall thickness and bronchiolar epithelial damage (Fig. 2 C). Moreover, Uhrf1MKO mice produced significantly more IFN-β in the serum than WT mice (Fig. 2 D). The elevated IFN-β induction contributed to H5N1 resistance in Uhrf1MKO mice, as suggested by the lower viral load in the lungs of these mice than in those of the WT control (Fig. 2 E). Additionally, Uhrf1MKO mice were more resistant to H1N1 infection than the control littermates, as indicated by a reduced body weight loss (Fig. 2 F) and an increased survival rate (Fig. 2 G). H&E staining showed that the infiltration of immune cells and injury to the lungs were lower in Uhrf1MKO mice than in WT mice after H1N1 infection (Fig. 2 H). Consistent with the increased production of IFN-β (Fig. 2 I), we also observed lower vesicular stomatitis virus (VSV) titers in the lungs of Uhrf1MKO mice (Fig. 2 J). In addition to RNA viruses, Uhrf1 deletion in myeloid cells also resulted in a decrease in the susceptibility to HSV-1 infection (Fig. 2, K and L) and enhancement of IFN-β production in the serum (Fig. 2 M). In the Ifnar1−/− background, Uhrf1MKO mice exhibited a survival rate and viral titer comparable to those of their littermates (Fig. 2, N and O), although they exhibited increased production of IFN-β (Fig. 2 P). Consistent with the observation in vivo, Uhrf1 deficiency in BM-derived macrophages (BMDMs) or MEFs significantly promoted WT or GFP-tagged VSV replication, as determined by plaque assays and flow cytometry analyses (Fig. 2, Q and R; and Fig. S2, A and B).
Figure S1.
Uhrf1 did not affect the development and homeostasis of the immune system. (A) Genotyping PCR of myeloid cell-conditional Uhrf1 WT (Uhrf1+/+lyz2Cre/+), Uhrf1MKO (Uhrf1fl/fllyz2Cre/+), and heterozygous (Uhrf1fl/+lyz2Cre/+) mice to amplify WT and floxed alleles and Cre-specific primers for the Lyz2Cre DNA. (B and C) Flow cytometry analysis of macrophages (CD11b+F4/80+) and neutrophils (CD11b+GR-1+) in BM and spleen (Spl) from 6–8-wk-old WT and Uhrf1MKO mice (n = 3). (D–G) Flow cytometry analysis of the absolute numbers of different immune cells in the spleen (D and E) and naive and memory T cells (F and G) from WT or USP16MKO mice (n = 3). All FACS data are presented in a representative plot and summary graph of the subpopulation percentages. All data are representative of three independent experiments. The bars and error bars show the means ± SEMs. Significance was determined by a two-tailed Student’s t test. BMC, BM cells; CM, central memory; EM, effector memory; iLN, inguinal LN; M, total memory; N, naïve; SP, splenocytes.
Figure 2.
Uhrf1 deficiency potentiates the antiviral immune response. WT and Uhrf1MKO mice (6–8 wk) were i.n. infected with a sublethal dose (0.1 HA) of H5N1 influenza virus. (A and B) The body weight loss (A) and survival rate (B) were measured for 14 d (n = 20). (C) H&E staining of lung tissue sections on days 2 and 5 after infection. Inflammation scores are presented as a bar graph (n = 5). Scale bar, 200 µm. (D) ELISA for IFN-β in the sera of WT and Uhrf1MKO mice infected with H5N1 influenza virus on days 2 and 5 (n = 3). (E) The viral titers in the lung were quantified on day 2 using the TCID50 assay (n = 10). (F–J) WT and Uhrf1MKO mice (6–8 wk) were i.n. infected with a sublethal dose (0.1 HA) of PR8. The body weight loss (F) and survival rate (G) were measured for 14 d (n = 12). H&E staining of lung tissue sections (H; n = 5) and IFN-β in serum (I; n = 4) was measured on days 2 and 5 after infection. Scale bars, 200 µm. (J) The viral titers in the lung were quantified on day 2 using the TCID50 assay (n = 10). (K–M) Survival rate (K; n = 12), viral titer (L; n = 8), and IFN-β in serum (M; n = 4) of WT and Uhrf1MKO mice intravenously injected with HSV-1 (3 × 106 PFU per mouse). (N–P) WT and Uhrf1MKO mice bred to the Ifnar1−/− background were i.n. infected with a sublethal dose (0.1 HA) of PR8. The survival rate (N; n = 16), viral titer (O; n = 5), and IFN-β in serum (P; n = 4) were monitored for 14 d. WT or Uhrf1-deficient BMDMs were infected with GFP-expressing VSV (VSV-GFP) at an MOI of 0.1 for 24 h. (Q) The data are presented as a representative picture, showing the infected (GFP+) and total (bright-field) cells. Scale bar, 1,000 µm. (R) Summary graph of flow cytometric quantification of the infected cells. All the data are representative of at least three independent experiments. The data are presented as means ± SEMs. The significance of differences was determined by a t test. *, P < 0.05; **, P < 0.01; ***, P < 0.005.
Figure S2.
Uhrf1 deficiency in myeloid cells caused an autoimmune feature. (A) Uhrf1ER-Cre MEFs were incubated with DMSO or 4-OH for 72 h and then infected with VSV-GFP at a MOI of 0.1 for 24 h Data are presented as a representative picture, showing the infected (GFP+) and total (bright-field) cells. Scale bar, 200 µm (n = 3). (B) The summary graph of flow cytometric quantification of the infected cells (n = 3). (C) Immunofluorescence microscopy of the deposition of IgM and IgG in glomeruli (arrows) of kidney sections from 8-mo-old WT and Uhrf1MKO mice. Original magnification, ×10. Scale bar, 1,000 µm. (D) ELISA of the autoantibodies antinuclear antigen (ANA) and antibody to double-stranded DNA (dsDNA) in the serum of these aging WT and Uhrf1MKO mice (n = 4). (E) Flow cytometric analysis of the percentages of IFN-γ– and IL-17–producing CD4+ T cells in the spleens and inguinal LNs (iLNs) of WT and Uhrf1MKO mice (n = 4). All FACS data are presented in a representative plot and summary graph of the subpopulation percentages. All data are representative of three independent experiments. The bars and error bars show the means ± SEMs. Significance was determined by two-tailed Student’s t test. *, P < 0.05, **, P < 0.01; ***, P < 0.005.
In addition to its role in the antiviral host defense, IFN-I functions as a central component in the pathological symptoms of systemic autoimmune diseases, such as systemic lupus erythematosus (SLE; Crow et al., 2019; Niewold, 2014). To evaluate the possible role of Uhrf1 in SLE pathogenesis, we evaluated kidney damage by tissue H&E staining and antibody deposition in kidney glomeruli (Fig. S2, C and D), which is a pathological feature of nephropathy. Uhrf1 deficiency in myeloid cells significantly enhanced the number of IFN-γ–producing T effector cells in the spleens of aging mice (Fig. S2, E and F). Overall, these results suggest that Uhrf1 plays a negative role in regulating host defense and antiviral innate immunity in vivo.
Uhrf1 deficiency promotes IFN-I production caused by viruses and TLR agonists
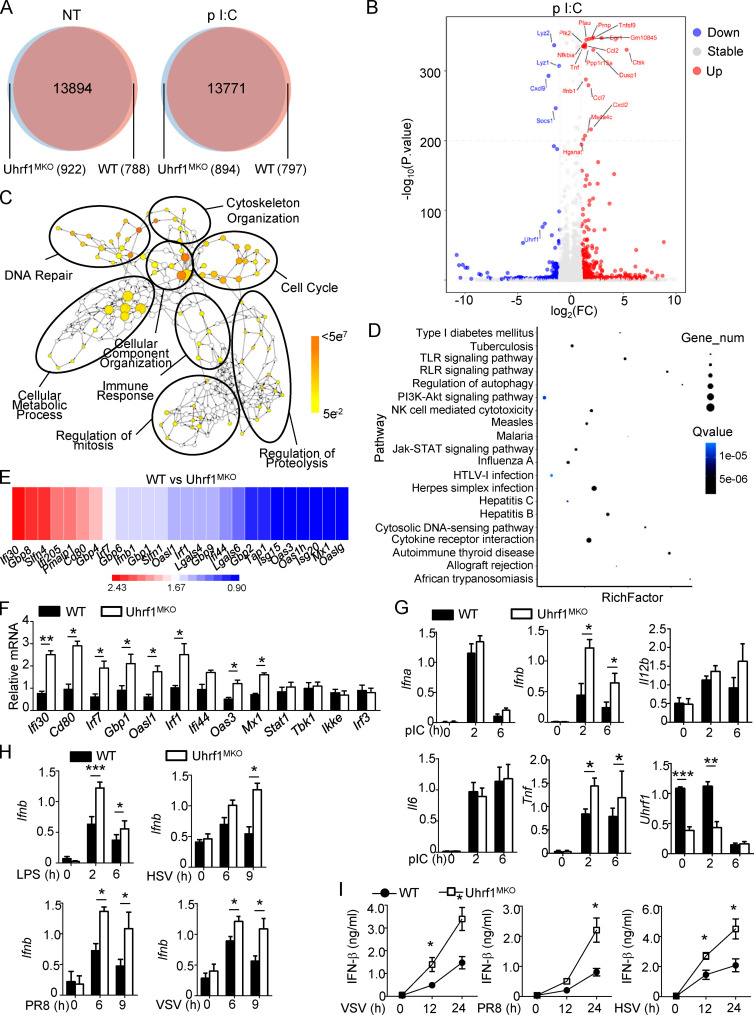
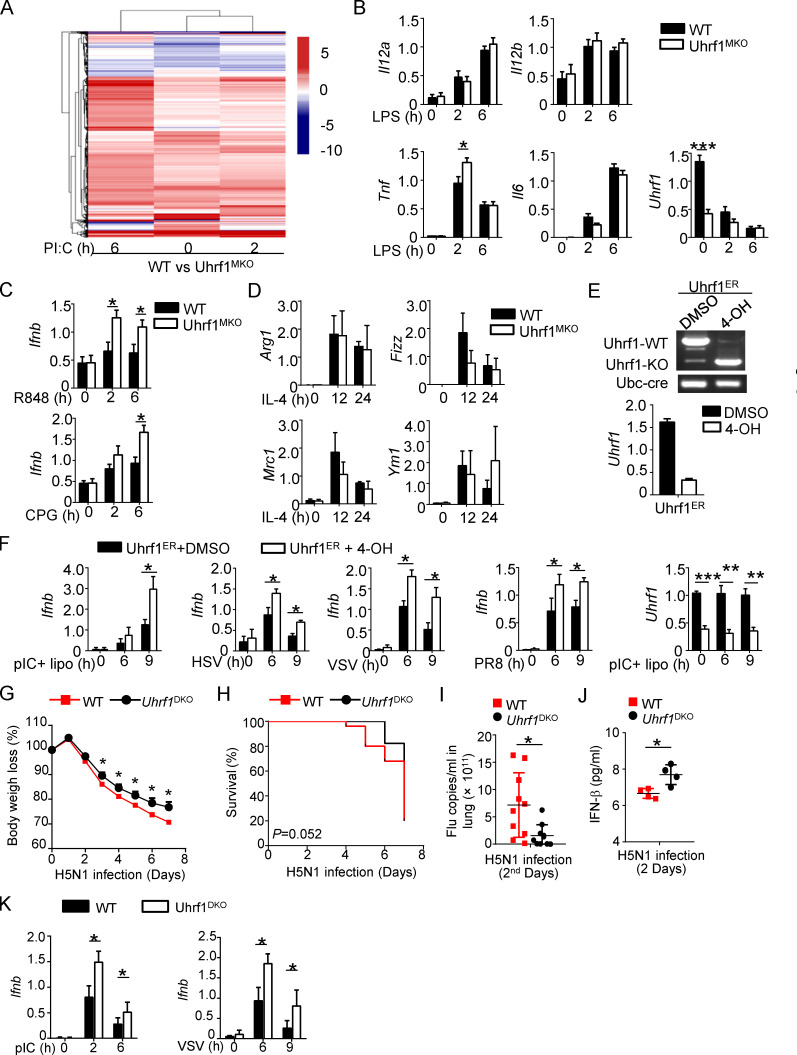
To further clarify the underlying mechanism by which Uhrf1 regulates the antiviral response, we monitored the effects of Uhrf1 on gene expression following poly(I:C) (pI:C) stimulation. Surprisingly, we only identified <1,000 differentially expressed genes (DEGs; more than twofold) in Uhrf1-deficient BMDMs than in the WT control (Fig. 3 A, Fig. S3 A, Table S1, and Table S2). The volcano plot and Ingenuity Pathway Analysis showed that several DEGs with altered major biological processes in Uhrf1-deficient BMDMs were enriched in multiple infectious diseases (Fig. 3, B and C). Gene ontology enrichment analysis indicated that only a small portion of the upregulated genes was enriched in the immune response to pI:C (Fig. 3 D). The heatmap revealed that Uhrf1 regulating the antiviral response is mainly mediated by IFNs and ISGs, such as Ifnb, Ifi30, and Oasl1 (Fig. 3 E). Additionally, real-time quantitative PCR confirmed the different expression levels of these ISGs in BMDMs from WT and Uhrf1MKO mice (Fig. 3 F and Fig. S3 B). Uhrf1 deficiency significantly enhanced Ifnb and Tnf induction in response to pI:C or LPS stimulation (Fig. 3 G). Additionally, Uhrf1 was a common regulator of Ifnb, as suggested by the decreased production of Ifnb caused by LPS, R848, CpG2216, HSV-1, flu (PR8), and VSV (Fig. 3 H and Fig. S3 C). The same results were obtained by ELISA, which evaluated cytokine secretion (Fig. 3 I). Interestingly, Uhrf1 was not required to express specific markers in M2 polarization (Fig. S3 D).
Figure 3.
Uhrf1 deficiency causes specific upregulation of IFN-I and ISGs. (A and B) Venn diagrams (A) and volcano graphs (B) illustrating the overlap of DEGs between WT and Uhrf1-deficient BMDMs stimulated with 20 µg/ml pI:C for 2 h. FC, fold change; NT, nontreatment. (C) Network visualization of gene ontology enrichment analysis of DEGs in Uhrf-deficient BMDMs in response to pI:C for 2 h. (D) KEGG analysis of these DEGs in Uhrf1-deficient BMDMs indicated pathways that differed significantly (in abundance). RLR, RIG-I–like receptor. (E and F) Heat map (E) and qPCR analysis (F) showing ISG expression in Uhrf1-deficient cells activated by pI:C for 2 h (n = 3). (G) qRT-PCR analysis of the indicated genes using WT or Uhrf1-deficient BMDMs stimulated with pI:C (n = 6). (H) The mRNA levels of Ifnb induced by different viruses in WT and Uhrf1-deficient BMDMs were measured by qPCR (n = 6). (I) ELISAs of IFN-β in the supernatants of WT or Uhrf1-deficient BMDMs stimulated with specific viruses (n = 3). All the data are representative of at least three independent experiments. The data from the qPCR assay are presented as fold changes relative to the actin mRNA levels. The data are presented as means ± SEMs. The significance of differences was determined by a t test. *, P < 0.05; **, P < 0.01; ***, P < 0.005.
Figure S3.
Uhrf1 is not required for the proinflammatory cytokines production except Ifnb. (A) Heatmap showing the pI:C-stimulated DEGs in BMDMs from WT and Uhrf1MKO mice. The DEGs were identified with a fold change of experimental sample to nontreated control (ES/NT) >2.0 or <0.5. (B) qRT-PCR analysis of the indicated genes using WT or Uhrf1-deficient BMDMs stimulated with 100 ng/ml LPS (n = 6). (C) Ifnb mRNA in Uhrf1MKO BMDMs were measured by qPCR assay responding to 1 μg/ml R848 or 5 μM CpG (n = 4). (D) qRT-PCR analysis of the indicated genes using WT or Uhrf1-deficicent BMDMs stimulated with 10 ng/ml IL-4 (n = 3). (E) Uhrf1ER-Cre MEFs were incubated with DMSO or 4-OH for 72 h, and we examined the genotyping by amplifying WT and KO alleles. qRT-PCR analysis of the Uhrf1 mRNA level. (F) qRT-PCR analysis of the indicated genes using WT or Uhrf1-deficient MEFs generated as above stimulated with pI:C plus 4-OH (n = 4). (G–J) WT and DC-conditional Uhrf1 KO (Uhrf1DKO) mice (6–8 wk) were i.n. infected with a sublethal dose (0.1 HA) of H5N1 influenza virus. Body weight loss (G) and survival rate (H) were measured for 14 d (n = 17). (I) Viral titers in the lung were quantified on day 2 by a TCID50 assay (n = 10). (J) ELISA for IFN-β in the sera of WT and Uhrf1MKO mice infected with H5N1 influenza virus on days 2 and 5 (n = 4). (K) qRT-PCR analysis of the indicated genes using WT or Uhrf1-deficient BM-derived DCs stimulated with 100 ng/ml LPS or 1 MOI VSV. Data in the qPCR assay are presented as fold relative to the actin mRNA level. All data are representative of at least three independent experiments. Data are presented as means ± SEMs. The significances of differences were determined by a t test. *, P < 0.05; **, P < 0.01; ***, P < 0.005.
To further confirm the essential role of Uhrf1 in promoting Ifnb production, Uhrf1ER MEFs were cultured with 4-OH before pI:C plus Lipofectamine stimulation (Fig. S3 E). Consistent with the observation in BMDMs, Uhrf1 deficiency also enhanced the induction of Ifnb in response to multiple virus infections (Fig. S3 F). To clarify the role of Uhrf1 in different cell types, we generated dendritic cell (DC)–conditional UHRF1-KO (Uhrf1DKO) mice by crossing Uhrf1flox mice with Itgaxcre/+ mice. These Uhrf1DKO mice also displayed modern resistance to flu infection, as suggested by lower body weight loss, death rate, and viral load (Fig. S3, G–I). There was also an increase in the induction of IFN-β in the serum of Uhrf1DKO mice (Fig. S3 J). Additionally, Uhrf1 deficiency in dendritic cells promoted the expression of Ifnb under pI:C or VSV stimulation (Fig. S3 K). Collectively, these data demonstrated that Uhrf1-mediated DNA methylation is involved in the expression of Ifnb in a common manner.
Regulation of IFN-I by Uhrf1 is independent of the canonical antiviral signaling pathway
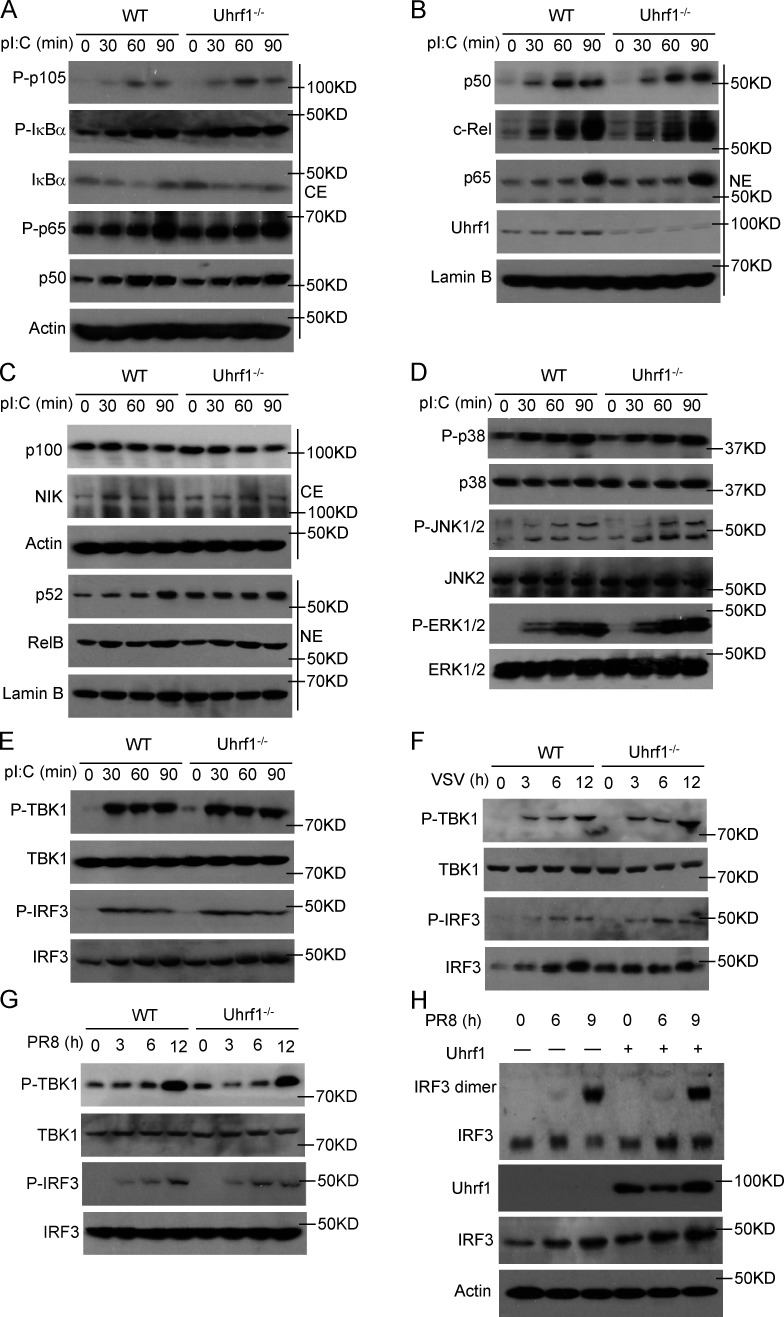
Recent evidence has revealed that p38 MAPK and canonical NF-κB activation are essential for IFN-I production and activation of myeloid innate cells (Bais et al., 2019; Mikkelsen et al., 2009; Wang et al., 2010); thus, we first examined canonical NF-κB activation. Immunoblot (IB) data indicated that Uhrf1 deficiency also did not significantly affect the activation of canonical NF-κB in BMDMs, as revealed by comparable levels of phosphorylated p105 and IΙκBα (Fig. 4 A). Additionally, Uhrf1 depletion was also dispensable for the nuclear translocation of three NF-κB members, c-Rel, p50, and p65 (Fig. 4 B). Our previous results demonstrated that the noncanonical NF-κB pathway negatively regulates the production of IFN-I during viral infection and suppresses antiviral host defense (Jin et al., 2014). However, the nuclear translocation of noncanonical NF-κB members, p52 and RelB, was not affected by Uhrf1 deficiency (Fig. 4 C). Furthermore, the hyperproduction of Ifnb was also not due to higher activation of the three major MAPK families in Uhrf1-deficient BMDMs (Fig. 4 D).
Figure 4.
Uhrf1 does not affect canonical antiviral signal transduction. (A–E) The indicated proteins in cytoplasmic (CE) and nuclear (NE) extracts (A–C) or whole-cell lysates (D and E) of WT and Uhrf1-deficient BMDMs were measured by IB analysis. (F and G) Specific virus-induced phosphorylation of TBK1 and IRF3 in whole-cell lysates was measured by IB analysis. (H) IB analysis of monomeric and dimeric IRF3 (top blot), total IRF3, Uhrf1, and actin (bottom) in HEK293T cells transfected with empty vector or an expression plasmid for Uhrf1 and then infected with PR8 for various times. IB analysis of monomeric and dimeric IRF3 (top blot), total IRF3, Uhrf1, and actin (bottom) in HEK293T cells transfected with empty vector or an expression plasmid for Uhrf1 and then infected with PR8 for various times. All the data are representative of at least three independent experiments.
Next, we examined the effect of Uhrf1 on the regulation of TLR- or virus-induced IFN-I signaling activation. In contrast to the higher Ifnb expression, Uhrf1 deficiency did not inhibit LPS-, VSV-, or PR8-induced phosphorylation of TBK1 and IRF3 (Fig. 4, E–G). Accordingly, PR8-induced IRF3 dimerization was not obviously different among Uhrf1-transfected HEK293T cells (Fig. 4 H). Collectively, our data suggested that Uhrf1 suppressed RNA virus–induced IFN-I production without affecting NF-κB, MAPK, or TBK1-IRF3 activation.
Uhrf1 deficiency results in single-nucleotide demethylation of the Ifnb promoter
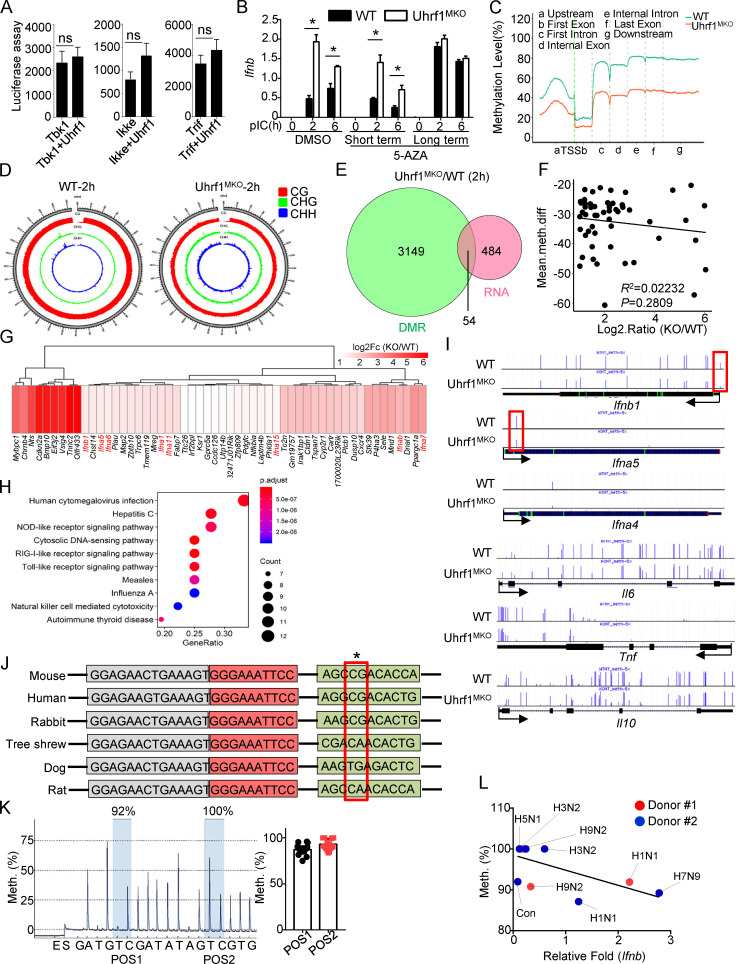
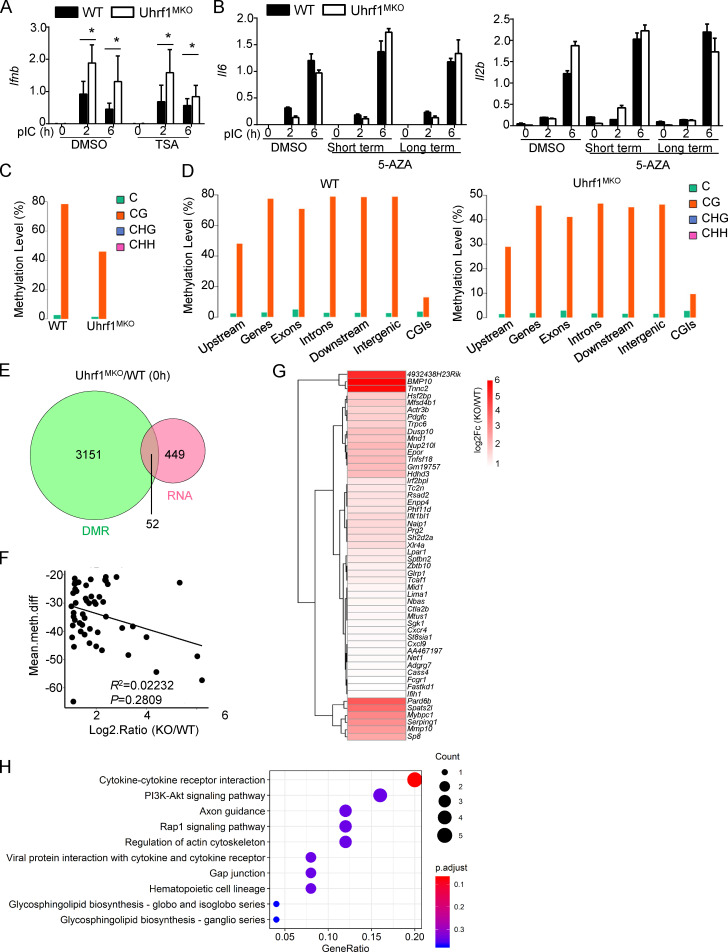
To further clarify the mechanism underlying Uhrf1 in the regulation of Ifnb induction, we examined the effect of Uhrf1 on Ifnb promoter activity via luciferase reporter assays in HEK293T cells. The overexpression of Uhrf1 did not lead to the inhibition of Ifnb promoter activation caused by TBK1, IKKe, or TRIF (Fig. 5 A). Uhrf1 regulates gene expression by recruiting the transcriptional repressors DNMT1 and HDAC1 through distinct domains. To examine whether HDAC1 is involved in Ifnb induction, we pretreated BMDMs with the HDAC inhibitor trichostatin A (TSA) before pI:C stimulation. The quantitative PCR (qPCR) data revealed that TSA treatment did not enhance Ifnb production (Fig. S4 A). We next tested whether DNA methylation is involved in Ifnb enhancement. WT and Uhrf1-deficient macrophages were preincubated with the DNA methylation inhibitor 5-azacytidine (5-AZA) before pI:C stimulation. We added 5-AZA at different time points for short-term incubation (day 4 during macrophage differentiation) and long-term incubation (day 0 during macrophage differentiation). As shown in Fig. 5 B, 5-AZA erased the differences in Ifnb production between WT and Uhrf1-deficient macrophages without affecting other proinflammatory cytokines (Fig. S4 B). These data suggest that the regulation of Ifnb in Uhrf1-deficient macrophages is mainly mediated by DNA methylation.
Figure 5.
Uhrf1 deficiency removes the methylation on the Ifnb promoter. (A) HEK293T cells were transfected with an Ifnb-luciferase reporter plasmid in the presence or absence of the indicated Uhrf1 expression plasmids (n = 4). Luciferase assays were performed, and the data are expressed as fold changes based on the empty vector group 36 h after transfection. (B) BM cells were incubated with 1 µM 5-AZA for 5 d (long). Differentiated macrophages were incubated with 1 µM 5-AZA for 24 h (short). The mRNA levels of Ifnb induced by pI:C in 5-AZA–pretreated WT and Uhrf1-deficient BMDMs were measured by qPCR (n = 5). (C) Methylation level (percentage) in the genomes of WT and Uhrf1-deficient BMDMs with the gene features of interest, including the gene body (all exons and introns), promoter, first exon, first intron, the rest of the exons, and the rest of the introns. (D) Epigenome density plot for CG, CHG, and CHH methylation contexts in chromosome 4 from WT and Uhrf1-deficient BMDMs. (E and F) Venn diagrams illustrating the overlap of DEGs whose promoters exhibited significant demethylation in pI:C-stimulated Uhrf1-deficient BMDMs (E), and correlation between the expression of these DEGs and methylation level on their promoter (F). (G and H) Heat map and KEGG analysis of these overlapping DEGs selected as above. (I) Genome Browser snapshots of the DNA methylation levels near Ifnb and other proinflammatory cytokine promoters in mice. (J) Sequence alignment of single-nucleotide methylated sites on Ifnb promoters of different species. The red box indicates the methylated site. (K) Pyrosequencing graphic results of the methylation levels of the Ifnb promoter region. The ratio of thymine (T) to cytosine (C) at each CpG position (gray) to determine the percentage of DNA methylation (n = 17). (L) Correlation between Ifnb and the methylation level on its promoter in PBMCs from healthy donors infected with different strains of the influenza virus. The data from the qPCR assay are presented as fold changes relative to the actin mRNA levels. All the data are representative of at least three independent experiments. The data are presented as means ± SEMs. The significance of differences was determined by a t test. *, P < 0.05.
Figure S4.
The transcriptome caused by Uhrf1 deficiency under nontreated conditions. (A) BMDMs were incubated with 10 μM TSA for 24 h. mRNA levels of Ifnb induced by pI:C WT and Uhrf1-deficient BMDMs were mentored by qPCR (n = 5). (B) BM cells were incubated with 1 μM 5-AZA for 5 d (long). Differentiated macrophages were incubated with 1 μM 5-AZA for 24 h (short). mRNA levels of Il6 and Il12b induced by pI:C in 5-AZA–pretreated WT and Uhrf1-deficient BMDMs were mentored by qPCR (n = 5). (C and D) Epigenome density plot for CG, CHG, and CHH methylation contexts in global genomes the genes feature of interest in WT and Uhrf1-deficient BMDMs, including the gene body (all exons and introns), promoter, first exon, first intron, the rest of the exons, and the rest of the introns. (E and F) Venn diagrams illustrating the overlap of DEGs whose promoter performed a significant demethylation in nontreated Uhrf1-deficient BMDMs (E). Correlation between these DEGs expression and methylation level on their promoter (F). (G and H) Heat map and KEGG analysis of these overlapped DEGs selected as above. Data in the qPCR assay are presented as fold relative to the actin mRNA level. All data are representative of at least three independent experiments. Data are presented as means ± SEMs. The significances of differences were determined by a t test. *, P < 0.05.
To further test whether an association exists between DNA methylation and Uhrf1-mediated gene expression, we first calculated the distribution of cytosine methylation levels. All the coding gene sequences were divided into seven different transcription element regions, and the methylation levels of these regions were calculated. The distribution characteristics of the DNA methylation levels revealed that Uhrf1 deficiency resulted in a global reduction in methylation at the whole-genome level (Fig. 5 C). Ifnb1 (ENSMUST00000055671.4) is located on chromosome 4, in which the methylation level is ∼80% for CG sites (Fig. 5 D and Fig. S4 C). Additionally, the distribution of these DNA methylation sites showed no preferred region (Fig. S4 D). DNA methylation in the promoter region is a repressive epigenetic modification that inhibits gene expression (Razin and Cedar, 1991; Schlosberg et al., 2017). Therefore, we further examined the association between Uhrf1 deficiency–mediated transcriptional regulation (with RNA sequencing [RNA-seq]) and DNA methylation in the promoter region. After pI:C stimulation, we observed that only a small portion of gene promoters exhibited a lower methylation level in Uhrf1-deficient BMDMs than in WT control BMDMs (Fig. 5 E). However, the methylation abundance in the promoter region was not correlated with the fold changes in these DEGs (Fig. 5 F). Consistent with the in vitro observations, Uhrf1-deficient BMDMs exhibited a significant upregulation of several IFN-I genes, including Ifnb and Ifna1/5/6/11/15, whose promoter regions presented with less methylation than those of their WT littermates (Fig. 5 G). Ingenuity Pathway Analysis indicated that the major biological processes were enriched in virus- or sensor-induced signaling pathways in Uhrf1-deficient BMDMs (Fig. 5 H). In contrast to the results obtained from pI:C treatment, the DEGs whose promoters displayed low methylation were mainly enriched in cytokine–cytokine receptor interactions and metabolic-related pathways (Fig. S4, E–H).
We next analyzed the role of DNA methylation in the promoters of specific proinflammatory cytokines. The upregulation of Ifnb most likely resulted from the hypomethylation of a single site at the proximal promoter region in the absence of Uhrf1 (Fig. 5 I). Interestingly, DNA methylation did not contribute to the expression regulation of other common cytokines, such as Il6, Tnf, or Il10 (Fig. 5 I). Interestingly, this specific CG site is conserved among mice, humans, and rabbits; dogs and rats have different sequences (Fig. 5 J). To examine the methylation level in human cells, the conserved methylated GC sites on the Ifnb promoter were verified in 36 samples by pyrosequencing. Surprisingly, the mean methylation level of PBMCs from healthy donors was 92.9 ± 6.0 (Fig. 5 K). Consistent with IAV strain–driven distinct expression of Uhrf1, pyrosequencing data also revealed that H5N1 infection caused a low methylation level at these specific GC sites that was maintained during H1N1 (PR8) infection (Fig. 5 L; different from Fig. 1). The different IAV strains also exhibited a negative correlation between the methylation level on this single site and Ifnb production (Fig. 5 L). These findings demonstrate that Uhrf1 regulation of Ifnb induction depends on the methylation level at a specific site of the Ifnb proximal promoter.
Alteration of specific CpG methylation on the promoter regulates Ifnb induction
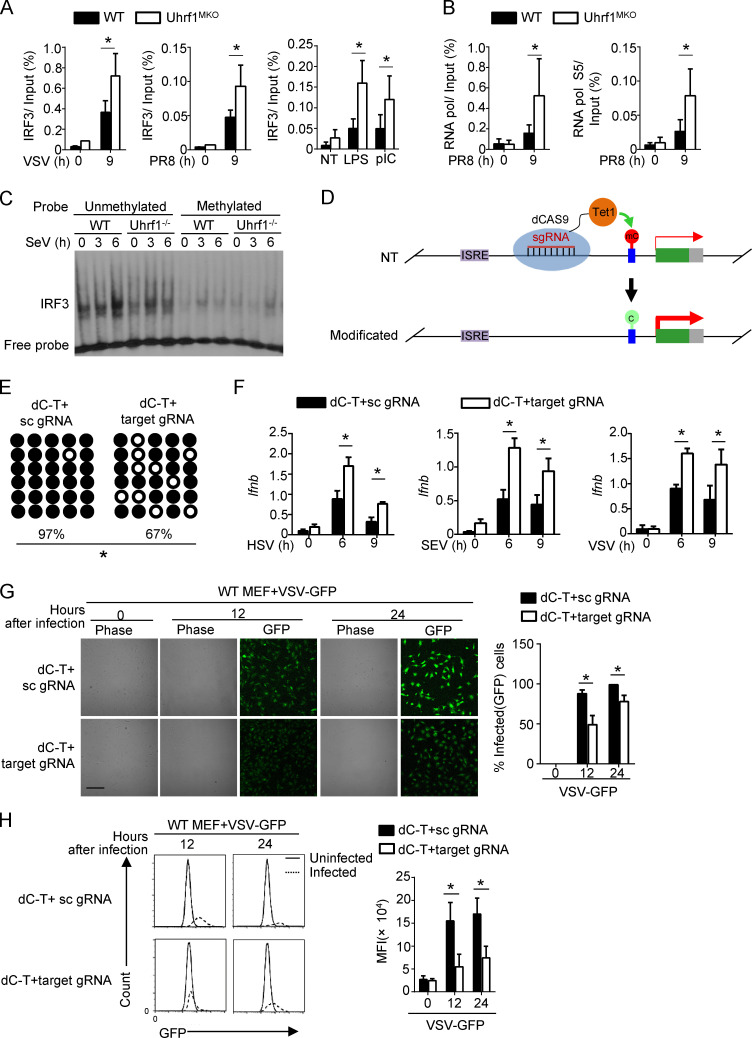
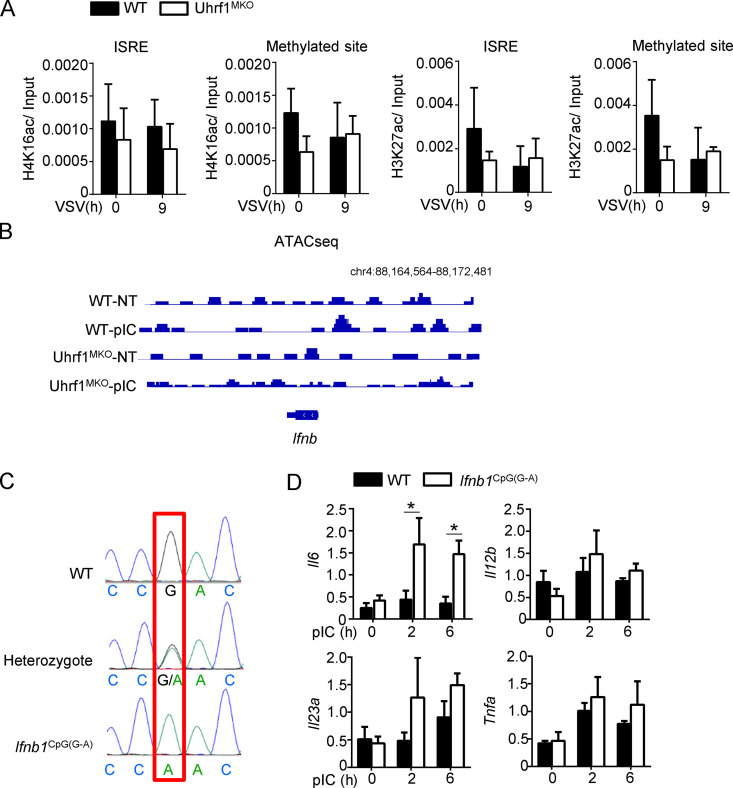
To elucidate the mechanism by which DNA methylation suppresses Ifnb induction, we evaluated the effect of Uhrf1 deficiency on the recruitment of IRF3 to the Ifnb promoter. Chromatin immunoprecipitation (ChIP) assays revealed that Uhrf1 deficiency promoted the recruitment of IRF3 to the Ifnb promoter after VSV, PR8, or TLR stimulation (Fig. 6 A). Ser5-phosphorylated RNA polymerase II is more concentrated at the promoter. Similar to IRF3, more Ser5-phosphorylated RNA polymerase II was recruited to the Ifnb promoter in response to PR8 infection than in response to WT control stimulation (Fig. 6 B). We next compared the ability of IRF3 to bind to the Ifnb methylated probe by performing the electrophoretic mobility-shift assay (EMSA). Uhrf1 deficiency did not affect IRF3 recruitment to the unmethylated probe. Moreover, this experiment indicated that the methylated probe significantly suppressed the binding of IRF3 (Fig. 6 C). K5, K8, K12, and K16 are frequently acetylated on histone H4. H4K16ac functions as a critical epigenetic mark involved in chromatin remodeling, DNA repair, and gene regulation (Samata et al., 2020; Urdinguio et al., 2019). Although a significant increase was found in H4K16ac in the monocytes of SLE patients (Leung et al., 2015), no difference was observed in the abundance of H4K16ac at the promoters of Ifnb when infected with VSV (Huai et al., 2019). Additionally, we found no high level or difference in H4K16ac on the Ifnb promoter between WT and Uhrf1-KO macrophages (Fig. S5 A). Additionally, H3K27ac is identified as an active marker on enhancers, indicating enhanced gene expression (Zhang et al., 2020). However, no correlation was found between Ifnb induction and H3K27ac. We also observed no enrichment of H3K27ac on the Ifnb promoter, and no difference was found between WT and Uhrf1-KO macrophages (Fig. S5 A). Next, we performed assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) to evaluate chromatin assembly near the Ifnb promoter (Table S3 and Table S4). No difference was found in chromatin assembly between the WT and Uhrf1-deficient groups under pI:C stimulation, suggesting that Uhrf1 did not regulate Ifnb expression via histone modification (Fig. S5 B).
Figure 6.
Targeting DNA methylation editing by dCas9-Tet1 activates IFN-I production. WT and Uhrf1-deficient BMDMs were stimulated for 9 h with specific virus or TLR agonists. (A and B) ChIP assays were performed and quantified by qPCR to detect the binding of IRF3 (A) or RNA polymerase (RNA pol; B) to the Ifnb promoter (n = 4). (C) EMSA of nuclear extracts of WT and Uhrf1-deficient BMDMs stimulated with Sendai Virus, as assessed using an unmethylated or methylated HRP-labeled Ifnb promoter probe. (D) Schematic graph of a catalytically inactive mutant Cas9 (dCas9) fused to Tet1 to remove DNA methylation modifications from the Ifnb promoter. (E) Bisulfite sequencing of WT MEFs transfected with dCas9-Tet1 (dC-T) plus a scrambled gRNA (sc gRNA) or four gRNAs targeting the Ifnb promoter region (target gRNA). (F) The mRNA levels of Ifnb induced by different viruses in MEFs described in E were measured by qPCR (n = 3). (G and H) MEFs described in B were infected with VSV-GFP at an MOI of 0.1 for 24 h. The data are presented as a representative picture, showing the infected (GFP+) and total (bright-field) cells (G; n = 3). Scale bar, 1,000 µm. Summary graph of flow cytometric quantification of the infected cells (H; n = 4). The data in the qPCR assay are presented as fold changes relative to the actin mRNA levels. All data are representative of at least three independent experiments. The data are presented as means ± SEMs. The significance of differences was determined by a t test. *, P < 0.05.
Figure S5.
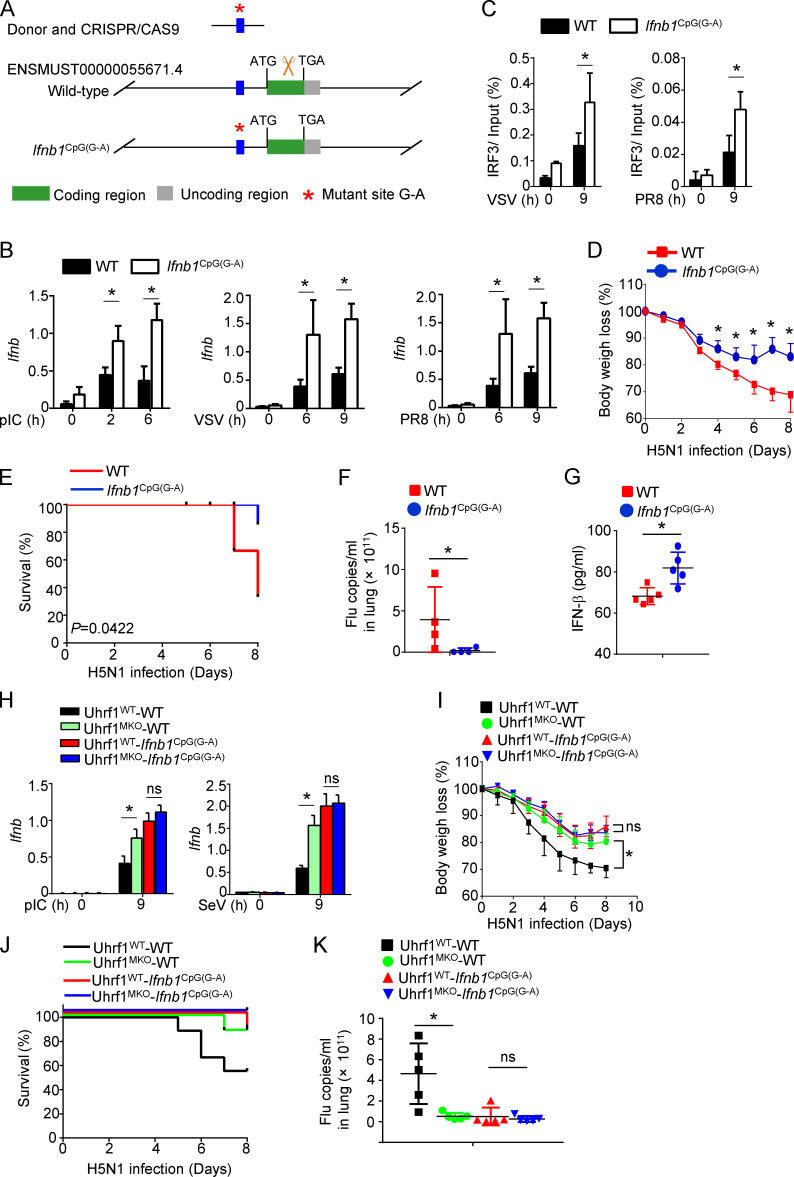
A single-nucleotide methylation negative regulated Ifnb induction independent of chromatin assembly. (A) WT and Uhrf1-deficient BMDMs were stimulated for 9 h with VSV. ChIP assays were performed and quantified by qPCR to detect the enrichment of H4K16ac and H3K27ac on the different regions of the Ifnb promoter (n = 4). (B) Screenshot from a genome browser of peaks near Ifnb obtained from ATAC-seq. (C) The sequencing data of WT and Ifnb1CpG(G-A) mice. (D) mRNA levels of indicated genes induced by 20 μg/ml pI:C in WT and Ifnb1CpG(G-A) BMDMs were mentored by qPCR (n = 4). Data in the qPCR assay are presented as fold relative to the actin mRNA level. All data are representative of at least three independent experiments. Data are presented as means ± SEMs. The significances of differences were determined by a t test. *, P < 0.05.
To generate a novel therapeutic tool for infectious disease based on DNA methylation, we fused dCas9 with Tet1 plus sequence-specific guide RNA (gRNA) to demethylate a specific locus without altering the DNA sequence (Fig. 6 D). Furthermore, bisulfite sequencing showed that the cotransfection of dCas9-Tet1 and gRNAs caused an obvious reduction in DNA methylation in the Ifnb promoter region (Fig. 6 E). To assess whether Ifnb induction could be activated by dCas9-Tet1 in WT MEFs, we transfected these MEFs with lentiviruses expressing dCas9-Tet1 with Ifnb promoter target gRNAs or a control gRNA. qPCR analyses revealed that only lentiviral vectors expressing dCas9-Tet1 with Ifnb promoter gRNAs, but not dCas9-Tet1 with control gRNA, promoted Ifnb production (Fig. 6 F). dCas9-Tet1 with Ifnb promoter gRNAs also suppressed GFP-tagged VSV replication in MEFs, as determined by plaque assays and flow cytometry analyses (Fig. 6, G and H).
Targeted demethylation of the Ifnb promoter enhances its transcription and the effect of the flu vaccine
We further generated a knock-in mouse model harboring a point mutation in the Ifnb promoter, designed to convert G to A (Ifnb1CpG(G-A)). To knock in this point mutation, we designed a single-guide RNA (sgRNA) targeting the Ifnb promoter carrying the intended substitution (Fig. 7 A and Fig. S5 C). As expected, the Ifnb1CpG(G-A) BMDMs were hyperresponsive to different stimulators in the induction of Ifnb (Fig. 7 B). In contrast to Uhrf1-deficient BMDMs, Ifnb1CpG(G-A) BMDMs exhibited increased IL-6 induction without affecting the production of other cytokines (Fig. S5 D). Additionally, the methylated point mutant enhanced the recruitment of IRF3 to the Ifnb promoter in response to VSV or PR8 (Fig. 7 C). Similar to the observation in vivo, less body weight loss and a lower IAV-induced mouse mortality rate were obtained in the Ifnb1CpG(G-A) mice than in the WT control (Fig. 7, D and E). The methylated point mutant resulted in a lower viral load in the lungs and a higher production of IFN-β in the serum than those in the WT littermates (Fig. 7, F and G). To confirm the role of this single-site methylation in Uhrf1 deficiency–mediated hyperproduction of Ifnb, we crossed Uhrf1MKO mice with Ifnb1CpG(G-A) mice. In Ifnb1CpG(G-A) background mice, a pivotal effect of Uhrf1 on IFN-I induction (Fig. 7 H) and antiviral response was significantly erased, as indicated by the comparable body weight loss, mortality rate, and number of flu copies in the lungs of the WT and Uhrf1MKO mice (Fig. 7, I and K). The data collectively suggest a negative role for Uhrf1-mediated site-specific methylation on the proximal promoter of Ifnb in regulating its production and expression in infectious diseases. The correlation between DNA methylation and Ifnb production suggests that the inhibitory effect of DNA methylation on the Ifnb promoter is a potential drug target for treating viral or bacterial infectious diseases.
Figure 7.
Ifnb production is inhibited by single-nucleotide methylation of its promoter. (A) Schematic picture showing the strategy for generating CpG mutant knock-in mice (Ifnb1CpG(G-A)). (B) The mRNA levels of Ifnb induced by different viruses in WT and Ifnb1CpG(G-A) BMDMs were measured by qPCR. ChIP assays were performed and quantified by qPCR to detect the binding of IRF3 to the WT or Ifnb1CpG(G-A) promoters (n = 4). (C) WT and Ifnb1CpG(G-A) mice (6–8 wk) were i.n. infected with a sublethal dose (0.1 HA) of H5N1 influenza virus (n = 4). (D and E) The body weight loss (D) and survival rate (E) were measured for 14 d (n = 18). (F) Viral titers in the lung were quantified on day 2 using the TCID50 assay (n = 4). (G) ELISA for IFN-β in the sera of WT and Ifnb1CpG(G-A) mice infected with H5N1 influenza virus on days 2 and 5 (n = 5). (H–K) WT and Uhrf1MKO mice bred on the Ifnb1CpG(G-A) background were i.n. infected with a sublethal dose (0.1 HA) of H5N1. The mRNA levels of Ifnb induced by different viruses in the indicated BMDMs were measured by qPCR (H; n = 3). The body weight loss (I; n = 8), survival rate (J; n = 8) for 14 d, and viral titer (K; n = 5) on day 2 are shown. The data in the qPCR assay are presented as fold changes relative to the actin mRNA levels. All data are representative of at least three independent experiments. The data are presented as means ± SEMs. The significance of differences was determined by a t test. *, P < 0.05.
Discussion
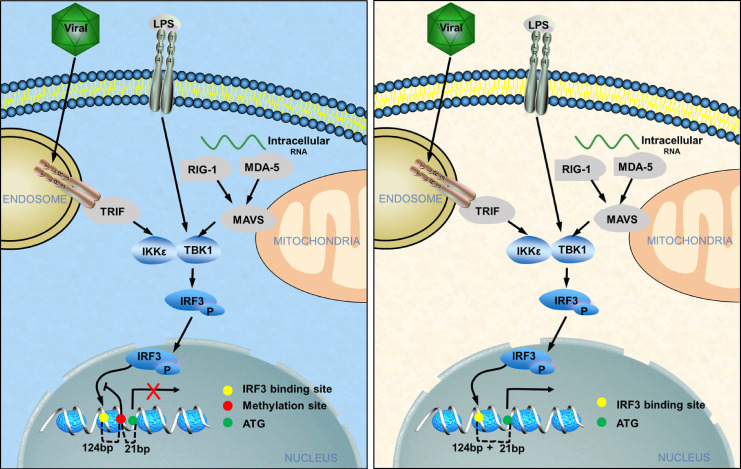
The occurrence of infectious diseases is closely related to the invasiveness of pathogens and host immune responses. Under normal conditions, innate immune cells, including macrophages and dendritic cells, are responsible for monitoring and maintaining the stability of the internal environment, preventing invasive pathogens and tissue damage, and removing these dangers. After virus infection, host innate immune cells trigger a protective mechanism that activates the antiviral signaling pathway by producing many IFN-Is to resist virus invasion. As a protective cytokine, IFN-I can eliminate the virus directly, subsequently promoting antigen-specific adaptive immunity. There are two main types of viruses, DNA and RNA viruses, which infect the target cell and trigger the TBK1-mediated IFN-I signaling pathway. DNA viruses activate STING by binding to intracellular cyclic GMP-AMP and further recruit the downstream kinase TBK1, while RNA viruses promote TBK1 by TLR-mediated signaling or recruiting it to mitochondrial antiviral signaling via RIG-I (Perry et al., 2005; Stetson and Medzhitov, 2006). When TBK1 is activated, it further phosphorylates IRF3 and triggers the translocation of the IRF3 homodimer from the cytoplasm to the nucleus. Nuclear IRF3 binds to the positive regulatory domain (PRD) region of the IFN-I promoter and facilitates the transcription of Ifna and Ifnb. Several studies have clarified how the TBK1-mediated pathway regulates virus infection and IFN-I production. However, it remains unclear whether epigenetic modifications, particularly DNA methylation, are involved in controlling the induction of IFN-I.
DNA methylation is a plastic process and plays pivotal roles in immune cell development and differentiation. DNA methylation can rapidly change to adapt to environmental perturbations, implying that DNA methylation is also highly involved in immune responses (Suarez-Alvarez et al., 2012). Generally, DNMT3A and DNMT3B add methylation modifications to the genome, and these modifications are further maintained by the complex of DNMT1 and Uhrf1/2 (Bostick et al., 2007; Razin and Cedar, 1991). Previous evidence has revealed that epigenetic modifications are involved in host antiviral host defense after IAV infection, and these changes in DNA methylation promote the expression of certain proinflammatory genes (Fang et al., 2012). Additionally, another study reported obvious changes in DNA methylation levels caused by different strains of IAV depending on their virulence (Mukherjee et al., 2013). Cells infected with the highly pathogenic H1N1 IAV displayed a significant reduction in DNA methylation levels on promoter regions. The new RNA virus COVID-19 is spreading quickly worldwide. COVID-19 infection causes a profound impairment of IFN-I production, as indicated by low IFN production and downregulation of ISGs (Hadjadj et al., 2020 Preprint). Current research has reported that DNA methylation is also involved in the expression of the COVID-19 host receptor ACE2 through genome-wide chromatin analysis (Corley and Ndhlovu, 2020 Preprint). These findings imply the significance of DNA methylation in understanding the interactions between viral infections and host immune defense.
The DNA methylation regulator UHRF1 is significantly correlated with immune infiltration in the tumor microenvironment. Additionally, our data revealed that distinct IAV strains resulted in different reductions in Uhrf1 expression, further contributing to the amount of Ifnb expressed. As an important epigenetic regulatory factor, UHRF1 contains several domains (ubiquitin-like, tandem tudor domain, plant homeodomain, SET- and RING-associated, and RING), each of which plays a role (Kong et al., 2019). UHRF1 has been reported to have important functions in immune system. In T cells, UHRF1 promotes the proliferation of colonic T reg cells in response to bacterial colonization by methylating the Cdkn1a promoter, which encodes the cyclin-dependent kinase inhibitor p21 (Obata et al., 2014). Similarly, Uhrf1 represses the expression of cdkn1a, slfn1, and slfn2 by DNA methylation and then facilitates GC B cell proliferation and affinity maturation (Chen et al., 2018). Additionally, Uhrf1 promotes invariant natural killer T cell development by regulating the Akt–mTOR signaling pathway (Cui et al., 2016). In macrophages, Uhrf1 inhibits Tnf expression by methylating the Tnf promoter to prevent inflammatory bowel diseases (Qi et al., 2019). However, our data revealed that Uhrf1-mediated Tnf expression is not directly regulated via DNA methylation. As known, UHRF1 functions as an essential partner in maintaining DNA methyltransferase with DNMT1, which directly methylates the newly synthesized daughter strand. Due to the defect of suitable ChIP antibody against UHRF1, we could not recognize which genes were specially regulated by UHRF1. However, whole-genome bisulfite sequencing data revealed that distinct motif preference of CG, CHG, and CHH. Therefore, the selective regulation for Ifnb by DNA methylation depends on the DNMT1-recognized specific sequence. The result of the defective methylation in Fig. 5 I indicated that Tnf, Il6, and Il10 could not be regulated by DNA methylation and demethylation. Surprisingly, whole-genome bisulfite sequencing analysis indicated a single and specific methylation site in the Ifnb promoter that directly contributed to Ifnb transcription.
5-AZA, a methyltransferase inhibitor, promotes IFN-I production. 5-AZA upregulates endogenous retrovirus transcripts through DNA demethylation and then induces the IFN-I response by triggering cytosolic sensing of double-stranded RNA (Chiappinelli et al., 2015; Topper et al., 2017). In contrast to treatment with methylation inhibitors, the signatures for IFN-α/β signaling were repressed in intrinsically DNA methylation–deficient tumors, likely reflecting IFN-α/β inactivation (Jung et al., 2019). Our data also did not show that basal Ifnb is upregulated in Uhrf1-deficient BMDMs, suggesting that 5-AZA–induced IFN-I is dependent on different cell types. Additionally, SLE is an autoimmune disease characterized by the hyperproduction of autoantibodies and IFN-I. Thus, we examined the hypothesis that the onset of SLE may be caused by disordered DNA demethylation on the promoter of Ifnb. Unfortunately, we found no hypomethylation of this region or a single-nucleotide mutant on the methylated site of the Ifnb promoter (data not shown), indicating that DNA methylation can be used to generate drugs to resist infectious diseases but does not contribute to the therapy of autoimmune diseases.
Dysregulation of DNA methylation leads to disordered gene regulation, resulting in multiple diseases (Wong and Wei, 2011). The relationship between DNA methylation and gene expression is complex because the modification occurs in different regions and specific cell types. DNA methylation regulates gene expression in various ways, including both positive (in gene body regions) and negative effects (in gene promoter regions). Here, we identified single-nucleotide methylation on the promoter of Ifnb that significantly suppressed Ifnb transcription. This single CpG methylation regulated Ifnb expression via the active mode, but not because of an independent mark to reflect the chromatin state. Our results indicated that this single CpG methylation modification disrupted the recruitment of IRF3 to the Ifnb promoter without affecting the chromatin accessibility characterized by ATAC-seq. This single CpG nucleotide is completely conserved between mice and humans but is different in rats or dogs. Interestingly, the IRF3 binding motif is 100 bp away from the CpG site; thus, methylation at this specific CpG site might not directly regulate IRF3 recruitment. The potential binding motifs of multiple TFs indicate that some TFs, including albumin D-binding protein, MyoG, MyoD, DREB1A, and Pax-6, may be regulators of IRF3 activity. A previous study demonstrated that GRIP1 acts as a negative regulator of MyoD-mediated transcription. GRIP1 knockdown in macrophages blocked IRF3-dependent gene expression, which was rescued by GRIP1 overexpression. This specific CpG site potentially disrupted the binding of these TFs to this motif, further inhibiting IRF3 recruitment to PRDIII and PRDI. It will be interesting to clarify how these TFs regulate DNA recognition in the future. These data indicate that the regulation of IFN-I may be different among species in response to the same pathogenic microorganisms.
In summary, we have identified single-CpG-nucleotide methylation as an essential mechanism that controls IFN-I induction and antiviral immunity in both humans and mice. By targeting this DNA methylation, we generated a novel adjuvant to induce specific demethylation of the Ifnb promoter, significantly enhancing the efficacy of the flu vaccine. Our study also provides functional insight into methylation loss in different IAV strain infections. We have provided genetic evidence that Uhrf1 expression is inhibited along with IAV infection, providing an example of how external stress regulates innate immunity epigenetically. Our findings also identified a potential function of single-nucleotide methylation in controlling IFN-I production. Our findings not only provide critical insight into the mechanism that regulates IFN-I production but also imply a potential drug target by modifying DNA methylation.
Materials and methods
Mice
Uhrf1fl/fl mice were produced by Shanghai Model Organisms Center. The Uhrf1-floxed mice were further crossed with Lyz2-Cre (004781) or ER-Cre (004682) mice (all from The Jackson Laboratory; C57BL/6 background) to generate macrophage conditional Uhrf1MKO (Uhrf1f/fLyz2-Cre) mice. Ifnb1 promoter mutant mice (Ifnb1CpG(G-A)) were generated by GemPharmatech. Ifnar1−/− mice were a gift from Bo Zhong (Wuhan University, Wuhan, China). Heterozygous mice were bred to generate littermate controls, and conditional KO mice were used for experiments. In the animal studies, the WT and multiple-KO mice were randomly grouped. Outcomes of animal experiments were collected blindly and recorded based on the ear-tag numbers of the experimental mice. The mice were maintained under specific pathogen–free conditions in a controlled environment of 50–70% humidity with a 12-h/12-h light/dark cycle and 20–22°C; food and water were provided ad libitum, and all animal experiments were conducted in accordance with protocols approved by the Institutional Animal Care and Use Committee of Zhejiang University. The gene-specific PCR primers used for genotyping are shown in Table S5.
Antibodies, plasmids, and reagents
Antibodies targeting IκBα (C-21, 1:1,000), p65 (C-20, 1:1,000), Lamin B (C-20, 1:1,000), ERK (K-23, 1:2,000), phospho-ERK (E-4, 1:1,000), JNK2 (C-17, 1:1,000), p38 (H-147, 1:1,000), p105/p50 (C-19, 1:1,000), TBK1 (108A429, 1:1,000), IRF3 (SC-9082, 1:1,000), RelB (C-19; 1:1,000), NIK (H248; 1:1,000), and c-Rel (sc-71, 1:1,000), as well as a control rabbit IgG (sc-2027), were obtained from Santa Cruz Biotechnology. Antibodies targeting phospho-IκBα (Ser32, 14D4, 1:1,000), phospho-JNK (Thr180/Tyr185, #9251, 1:1,000), phospho-p38 (Thr180/Tyr182, 3D7, 1:1,000), phospho-p105 (Ser933, 18E6, 1:1,000), phospho-TBK1 (Ser172, D52C2, 1:1,000), phospho-IRF3 (Ser396,4D4G, 1:1,000), Uhrf1 (D6G8E, 1:1,000), and p100/p52 (4882; 1:1,000) were purchased from Cell Signaling Technology. Anti-actin (C-4, 1:10,000) was from Sigma-Aldrich. Other fluorescence-labeled antibodies are listed in the flow cytometry and cell sorting sections.
dcas9-Tet1 and gRNA plasmids were provided by Li Shen (Zhejiang University, Hangzhou, China). pMD2. G and psPAX2 plasmids were provided by Yichuan Xiao (Chinese Academy of Sciences, Shanghai, China).
CpG (2216) was purchased from Sigma-Aldrich. R848 and pI:C were purchased from Amersham Biosciences. The inactivated virus vaccine VaxigripTetra (IIV4) was purchased from Sanofi Pasteur.
Flow cytometry and intracellular staining
Single-cell suspensions from the BM, spleen, or draining lymph nodes were subjected to flow cytometry using CytoFlex (Beckman Coulter) and the following fluorescence-labeled antibodies from eBioscience: Pacific Blue–conjugated anti-CD4 and anti-CD11c, PE-conjugated anti-F4/80 and anti–IL-17, PerCP5.5-conjugated anti–Gr-1 (Ly6G), APC-conjugated anti-CD62L, APC-Cy7–conjugated anti-CD11b and anti-CD8, and FITC-conjugated anti–IFN-γ and anti-CD44.
For intracellular cytokine staining, single-cell suspensions from spleens or draining lymph nodes were stimulated with PMA (0.5 µg/ml) plus ionomycin (1 µg/ml) for 3 h and monensin (eBioscience; ×1,000) for another 3 h, followed by intracellular IFN-γ and IL-17 by flow cytometry analysis. All FACS data were analyzed by FlowJo 7.6.1.
Generation of BMDMs
WT or Uhrf1MKO mice were sacrificed by cervical dislocation and the removal of both femurs. After removing the muscle and fat with a pair of scissors, the femurs were placed in 5 ml DMEM on ice. The ends of the bone were cut with a sharp pair of scissors in the tissue culture hood, and then the BM was flushed out with a 10-ml syringe (25-G needle) and DMEM. BM cells were collected in a 15-ml tube and centrifuged at 1,600 rpm for 5 min. BMDMs were cultured in DMEM supplemented with 20% FBS and 30% L929 cell-conditioned medium. The growth medium was changed on the third day. The cells were harvested using 2.5 mM EDTA prepared in PBS supplemented with 5% FBS on the fifth day and replated to obtain an even number of cells on different plates for further experiments.
BMDMs were starved overnight in medium supplemented with 1% FBS before stimulation with pI:C (20 µg/ml), CpG (2216, 5 µM), or R848 (1 µg/ml) for cytokine induction experiments. Total and subcellular extracts were prepared for IB assays, and total RNA was prepared for quantitative RT-PCR (qRT-PCR) assays.
EMSA
EMSA was performed as previously described (Maggirwar et al., 1997) using a biotin-labeled oligonucleotide probe covering the IFN-β NF-κB element (pRD1-II), and the specific methylated site shown in Table S6.
Isolation of PBMCs and MEFs
Anticoagulated blood samples from healthy donors (College of Animal Sciences, Zhejiang University, and Life Sciences Institute, Zhejiang University) were used to isolate PBMCs using lymphocyte separation medium (MULTISCIENCES) and density-gradient centrifugation. The use of PBMCs complied with the institutional guidelines and approved protocols of Zhejiang University.
For MEFs, heterozygous mice were bred to obtain Uhrf1ER-Cre and WT embryos from the same pregnant female mice. Embryos were removed from the mice at embryonic days 13.5–14.5 of gestation and then cut and enzymatically disaggregated after removing blood and liver tissue to generate single MEFs.
ELISA and real-time qRT-PCR
The supernatants of in vitro cell cultures were analyzed by ELISA using a commercial assay system (eBioscience). Autoantibodies for double-stranded DNA and nuclear antigen in the sera collected from 10-mo-old mice were measured using specific ELISA kits (Alpha Diagnostic).
For qRT-PCR, total RNA was isolated using TRIzol reagent (Molecular Research Center) and subjected to cDNA synthesis using RNase H-reverse transcription (Invitrogen) and oligo (dT) primers. qRT-PCR was performed in triplicate using the iCycler Sequence Detection System (Bio-Rad) and iQTM SYBR Green Supermix (Bio-Rad). The expression of individual genes was calculated using a standard curve method and normalized to the expression of Actb. The gene-specific PCR primers (all for mouse genes) are shown in Table S7.
ChIP assay
ChIP assays were performed using macrophages stimulated for 6 h with 20 µg/ml pI:C or MEFs stimulated for 9 h with pI:C plus Lipofectamine. The cells were fixed with 1% formaldehyde and sonicated as previously described (Nelson et al., 2006). Lysates were subjected to IP with the appropriate antibodies, and the precipitated DNA was then purified using Qiaquick columns (Qiagen) and quantified by qPCR using a pair of primers that amplifies the target region of the indicated promoter. The precipitated DNA is presented as the frequency of the total input DNA. For histone modification analyses, the DNA bound by modified histone H3 is presented as the frequency of total histone H3–bound DNA. The ChIP-qPCR primers are shown in Table S7.
Preparation of nuclear and cytosolic extracts and whole-cell lysates
Cells were lysed in buffer B lysis buffer (10 mM Hepes, pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.4% NP-40, 1 mM dithiothreitol [DTT], and 0.1 mM PMSF) and incubated on ice for 15 min. After spinning (12,000 rpm) for 15 s, the supernatant was transferred to a new tube (cytosolic extract). The nuclear pellets were washed with 500 µl buffer B three times. After removing the washing buffer, buffer C (20 mM Hepes, pH 7.9, 0.4 mM NaCl, 1 mM EDTA, 1 mM DTT, and 1 mM PMSF) was added to the nuclear pellets. These pellets were shaken vigorously for 10 min on a vortex shaker (4°C). Following centrifugation (12,000 rpm) for 1 min, the supernatants were collected and transferred to a new tube (nuclear extract). β-Actin and Lamin B were used as loading controls for the cytoplasm and nucleus, respectively. Whole-cell lysates were lysed in RIPA buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% NP-40, 0.5% Na-deoxycholate, 1 mM EDTA, 1 mM DTT, and 1 mM PMSF) on ice for 15 min, and the supernatants were collected after centrifugation.
Virus infection
Age-matched (6–8 wk old) mice were housed in microisolation cages at a biosafety level 3 facility. The mice were anesthetized with dry ice, and the nasal cavity was dripped with 106 TCID50 H1N1 (PR8), H5N1, or H7N9 virus (obtained from the Center for Disease Control and Prevention of Zhejiang Province, Hangzhou, China) in 40 µl PBS. The infected mice were monitored for body weight and lethality for up to 14 d. Samples of sera, lung, and spleen from infected mice were collected immediately upon sacrifice on days 2 and 5. The sera were used to measure IFNs by ELISA, and the tissues were homogenized to determine the number of virous copies. Additionally, a fraction of the lung in the same area was placed in formalin and processed for histology. For in vitro virus infection, the cells were infected with H7N9 virus at a multiplicity of infection (MOI) of 1 at the indicated times in the figure legends. After adsorption for 1 h at 37°C, the cells were washed with PBS and cultured in DMEM containing 2 µg/ml L-1-tosylamido-2-phenylethyl chloromethyl ketone–treated trypsin. Virus stocks were propagated in embryonated chicken eggs, and the virus titer was measured as the TCID50 in MDCK cells. For the in vitro infections with VSV, Sendai Virus, or HSV-1, MOI = 1.
Western blotting
Whole-cell lysates or subcellular extracts were prepared and subjected to IB and IP assays as mentioned above. The samples were resolved by 8.25% SDS-PAGE. After electrophoresis, the separated proteins were transferred onto polyvinylidene difluoride membranes (Millipore). For IB, the polyvinylidene difluoride membrane was blocked with 5% nonfat milk. After incubation with a specific primary antibody, an HRP-conjugated secondary antibody was applied. The positive immune reactive signal was detected by enhanced chemiluminescence (Amersham Biosciences).
Histology and immunohistochemistry
The lungs from untreated or virus-infected mice were fixed in 4% (wt/vol) paraformaldehyde, embedded in paraffin, and sectioned for H&E staining. The slides were numbered randomly, and the quantity of perivascular or peribronchial inflammation was blindly analyzed using Image-Pro software.
For immunohistochemistry, the kidneys from 10-mo-old mice were immersed in optimal cutting temperature compound and frozen at −80°C until sectioning. Cryostat sections (6 µm) of kidneys were prepared and stained with Alexa Fluor 594–conjugated goat antibody to mouse IgG (Invitrogen; A-11005).
Fluorescence microscopy
MEFs were seeded into 12-well plates, infected with VSV-GFP in serum-free medium for 1 h, and replaced with growth medium for another 11 h. Cells treated with or without stimulation were fixed with 4% paraformaldehyde for 20 min. Next, the cells were washed with PBS three times. The infected (GFP+) cells were visualized under a fluorescence microscope and quantified by flow cytometry.
dCase9-Tet1 assay
dCas9-Tet1 mediates demethylation of the gene promoter as previously described (Nelson et al., 2006). dCas9-Tet1 and gRNA expression plasmids were transfected into MEFs through a lentiviral system using a standard transduction protocol, and the transduced cells were isolated using a FACS sorter (Aria II) based on GFP (gRNA expression plasmids carrying the GFP gene). The gRNA sequences targeting Ifnb were as follows: Ifnb sgRNA1, 5′-CCATCCCTTATAAATAGCACAGG-3′; Ifnb sgRNA2, 5′-GATCCACCTGTTGTTCATGATGG-3′.
Bisulfite sequencing analysis
DNA was extracted from MEFs transfected with dCas9-Tet1 using the DNeasy Blood & Tissue Kit (Qiagen; #69504) and then was subjected to bisulfite conversion using the EpiTect Bisulfite Kit (Qiagen; #59104). The PCR amplicons were cloned into the pMD18-T vector (TaKaRa), followed by sequencing. The primers were designed using MethPrimer software (http://www.urogene.org/methprimer2/) and listed as follows: 5′-AAATAGTATAGGTTATGAAGGAAGATTA-3′ (forward) and 5′-TCTTTCCATTCAACTACTCCAAAA-3′ (reverse).
RNA-seq analysis
The RNA samples were collected from WT and Uhrf1 MKO BMDMs with or without pI:C stimulation. Total RNA was isolated and reverse transcribed into cDNA to generate an indexed Illumina library, followed by sequencing at the Beijing Genomics Institute (Beijing, China) using a BGISEQ-500 platform. High-quality reads were aligned to the mouse reference genome (mm10) using Bowtie2. The expression of individual genes was normalized to fragments per kilobase per million mapped reads from RNA-seq by expectation maximization. Significant differential expression was set if a gene showed a greater than onefold expression difference versus the control with adjusted P value of <0.05. The DEGs were analyzed by gene ontology using AMIGO and DAVID software. The enrichment degrees of DEGs were analyzed using Kyoto Encyclopedia of Genes and Genomes (KEGG) annotations.
Statistical analysis
Statistical analysis was performed using Prism software. No data were excluded from the analyses. A two-tailed unpaired t test was performed. P values <0.05 were considered statistically significant, and the levels of significance are indicated as *, P < 0.05 and **, P < 0.01. In the animal studies, at least four mice were required for each group based on the calculated number necessary to achieve a 2.3-fold change (effect size) in a two-tailed t test with 90% power and a significance level of 5%. All statistical tests were justified as appropriate, and the data met the assumptions of the tests. The variance was similar between the statistically compared groups.
Data availability
The raw sequence data reported in this paper were deposited in the Genome Sequence Archive (Wang et al., 2017) at the BIG Data Center (National Genomics Data Center Members and Partners, 2020), Beijing Institute of Genomics, Chinese Academy of Sciences, under the accession no. PRJCA002630.
Online supplemental material
Fig. S1 shows Uhrf1 deficiency in myeloid cells does not involved in the development of macrophage and neutrophils and the homeostatic of T lymphocytes. Fig. S2 shows Uhrf1 deficiency does not only promote antiviral response but also causes autoinflammation. Fig. S3 shows Uhrf1 negatively regulates the induction of Ifnb but is dispensable for the proinflammatory cytokines production. Fig. S4 shows the DEGs between WT and Uhrf1-deficient BMDMs without stimulation. Fig. S5 shows chromatin assembly does not contribute to the hyperproduction of Ifnb induced by Uhrf1 loss. Table S1 lists DEGs between WT and Uhrf1-deficient BMDMs without stimulation. Table S2 lists the DEGs between WT and Uhrf1-deficient BMDMs stimulated with 20 µg/ml pI:C for 2 h assessed by mRNA sequencing. Table S3 lists ATAC-seq of WT and Uhrf1-deficient BMDMs without stimulation. Table S4 lists ATAC-seq of WT and Uhrf1-deficient BMDMs under pI:C stimulation. Table S5 lists the sequences of genotyping primers. Table S6 lists the sequences of probes used for EMSA. Table S7 lists the oligonucleotide sequences used for qPCR.
Supplementary Material
lists DEGs between WT and Uhrf1-deficient BMDMs without stimulation.
lists the DEGs between WT and Uhrf1-deficient BMDMs stimulated with 20 µg/ml pI:C for 2 h assessed by mRNA sequencing.
lists ATAC-seq of WT and Uhrf1-deficient BMDMs without stimulation.
lists ATAC-seq of WT and Uhrf1-deficient BMDMs under pI:C stimulation.
lists the sequences of genotyping primers.
lists the sequences of probes used for EMSA.
lists the oligonucleotide sequences used for qPCR.
Acknowledgments
We thank the Life Sciences Institute core facilities, Zhejiang University, for technical assistance. We also thank Shao-cong Sun (MD Anderson Cancer Center, Houston, TX) for the expression plasmid.
This study was supported by the National Key Research and Development Program of China (2018YFD0500100 and 2018YFA0800503), the Excellent Young Scientist Fund of National Natural Science Foundation of China (31822017), the Natural Science Foundation of Zhejiang Province (LR19C080001), National Natural Science Foundation of China (81771675 and 81970484), and the Key Research and Development Program of Zhejiang Province (2020C03075).
Author contributions: Conceptualization, J. Jin and Y.-y. Li; Methodology, Z.-j. Gao, W.-p. Li, and X.-t. Mao; Software, X.-t. Mao; Investigation, Z.-j. Gao, W.-p. Li, T. Huang, H.-l. Wang, Y.-n. Li, and B.-q. Liu; Data curation, J.-y. Zhong; Writing – original draft, J. Jin, Y.-y. Li, and Z.-j. Gao; Writing – review and editing, J. Jin, Y.-y. Li, and C. Renjie; Visualization, J. Jin, Z.-j. Gao, W.-p. Li, and Y.-y. Li; Supervision, J. Jin and Y.-y. Li; Funding acquisition, J. Jin.
References
- Aune, T.M., Collins P.L., Collier S.P., Henderson M.A., and Chang S.. 2013. Epigenetic activation and silencing of the gene that encodes IFN-g. Front. Immunol. 4:112. 10.3389/fimmu.2013.00112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bais, S.S., Ratra Y., Khan N.A., Pandey R., Kushawaha P.K., Tomar S., Medigeshi G., Singh A., and Basak S.. 2019. Chandipura Virus Utilizes the Prosurvival Function of RelA NF-κB for Its Propagation. J. Virol. 93:e00081–19. 10.1128/JVI.00081-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylin, S.B., and Jones P.A.. 2016. Epigenetic Determinants of Cancer. Cold Spring Harb. Perspect. Biol. 8:a019505. 10.1101/cshperspect.a019505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommarito, P.A., and Fry R.C.. 2019. The Role of DNA Methylation in Gene Regulation. In Toxicoepigenetics. McCullough S.D., and Dolinoy D.C., editors. Academic Press, Cambridge. pp. 127–151. 10.1016/B978-0-12-812433-8.00005-8 [DOI] [Google Scholar]
- Bostick, M., Kim J.K., Estève P.-O., Clark A., Pradhan S., and Jacobsen S.E.. 2007. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 317:1760–1764. 10.1126/science.1147939 [DOI] [PubMed] [Google Scholar]
- Carty, S.A., Gohil M., Banks L.B., Cotton R.M., Johnson M.E., Stelekati E., Wells A.D., Wherry E.J., Koretzky G.A., and Jordan M.S.. 2018. The Loss of TET2 Promotes CD8+ T Cell Memory Differentiation. J. Immunol. 200:82–91. 10.4049/jimmunol.1700559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C., Zhai S., Zhang L., Chen J., Long X., Qin J., Li J., Huo R., and Wang X.. 2018. Uhrf1 regulates germinal center B cell expansion and affinity maturation to control viral infection. J. Exp. Med. 215:1437–1448. 10.1084/jem.20171815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernyavskaya, Y., Mudbhary R., Zhang C., Tokarz D., Jacob V., Gopinath S., Sun X., Wang S., Magnani E., Madakashira B.P., et al. 2017. Loss of DNA methylation in zebrafish embryos activates retrotransposons to trigger antiviral signaling. Development. 144:2925–2939. 10.1242/dev.147629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiappinelli, K.B., Strissel P.L., Desrichard A., Li H., Henke C., Akman B., Hein A., Rote N.S., Cope L.M., Snyder A., et al. 2015. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via dsRNA Including Endogenous Retroviruses. Cell. 162:974–986. 10.1016/j.cell.2015.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corley, M.J., and Ndhlovu L.C.. 2020. DNA Methylation Analysis of the COVID-19 Host Cell Receptor, Angiotensin I Converting Enzyme 2 Gene (ACE2) in the Respiratory System Reveal Age and Gender Differences. Preprints. (Preprint posted March 19, 2020) 10.20944/preprints202003.0295.v1 [DOI]
- Crow, M.K., Olferiev M., and Kirou K.A.. 2019. Type I Interferons in Autoimmune Disease. Annu. Rev. Pathol. 14:369–393. 10.1146/annurev-pathol-020117-043952 [DOI] [PubMed] [Google Scholar]
- Cui, Y., Chen X., Zhang J., Sun X., Liu H., Bai L., Xu C., and Liu X.. 2016. Uhrf1 Controls iNKT Cell Survival and Differentiation through the Akt-mTOR Axis. Cell Rep. 15:256–263. 10.1016/j.celrep.2016.03.016 [DOI] [PubMed] [Google Scholar]
- Cull, A., Snetsinger B., and Rauh M.J.. 2015. Tet2 Is a Novel Regulator of Murine Macrophage Differentiation and Polarization. Blood. 126:646. 10.1182/blood.V126.23.646.646 [DOI] [Google Scholar]
- Cull, A.H., Snetsinger B., Buckstein R., Wells R.A., and Rauh M.J.. 2017. Tet2 restrains inflammatory gene expression in macrophages. Exp. Hematol. 55:56–70.e13. 10.1016/j.exphem.2017.08.001 [DOI] [PubMed] [Google Scholar]
- Davis, F.M., and Gallagher K.A.. 2019. Epigenetic Mechanisms in Monocytes/Macrophages Regulate Inflammation in Cardiometabolic and Vascular Disease. Arterioscler. Thromb. Vasc. Biol. 39:623–634. 10.1161/ATVBAHA.118.312135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, Y., Mo X., Hu Y., Qi X., Jiang F., Jiang Z., and Tong S.. 2020. Epidemiology of COVID-19 Among Children in China. Pediatrics. 145:e20200702. 10.1542/peds.2020-0702 [DOI] [PubMed] [Google Scholar]
- Fang, J., Hao Q., Liu L., Li Y., Wu J., Huo X., and Zhu Y.. 2012. Epigenetic changes mediated by microRNA miR29 activate cyclooxygenase 2 and lambda-1 interferon production during viral infection. J. Virol. 86:1010–1020. 10.1128/JVI.06169-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamper, C.J., Agoston A.T., Nelson W.G., and Powell J.D.. 2009. Identification of DNA methyltransferase 3a as a T cell receptor-induced regulator of Th1 and Th2 differentiation. J. Immunol. 183:2267–2276. 10.4049/jimmunol.0802960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjadj, J., Yatim N., Barnabei L., Corneau A., Boussier J., Pere H., Charbit B., Bondet V., Chenevier-Gobeaux C., Breillat P., et al. 2020. Impaired type I interferon activity and exacerbated inflammatory responses in severe Covid-19 patients. medRxiv. (Preprint posted April 23, 2020) 10.1101/2020.04.19.20068015 [DOI] [PMC free article] [PubMed]
- He, Y.-F., Li B.-Z., Li Z., Liu P., Wang Y., Tang Q., Ding J., Jia Y., Chen Z., Li L., et al. 2011. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 333:1303–1307. 10.1126/science.1210944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotchkiss, R.D. 1948. The quantitative separation of purines, pyrimidines, and nucleosides by paper chromatography. J. Biol. Chem. 175:315–332. 10.1016/S0021-9258(18)57261-6 [DOI] [PubMed] [Google Scholar]
- Huai, W., Liu X., Wang C., Zhang Y., Chen X., Chen X., Xu S., Thomas T., Li N., and Cao X.. 2019. KAT8 selectively inhibits antiviral immunity by acetylating IRF3. J. Exp. Med. 216:772–785. 10.1084/jem.20181773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichiyama, K., Chen T., Wang X., Yan X., Kim B.-S., Tanaka S., Ndiaye-Lobry D., Deng Y., Zou Y., Zheng P., et al. 2015. The methylcytosine dioxygenase Tet2 promotes DNA demethylation and activation of cytokine gene expression in T cells. Immunity. 42:613–626. 10.1016/j.immuni.2015.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa, J.-P.J., and Kantarjian H.M.. 2009. Targeting DNA methylation. Clin. Cancer Res. 15:3938–3946. 10.1158/1078-0432.CCR-08-2783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, J., Hu H., Li H.S., Yu J., Xiao Y., Brittain G.C., Zou Q., Cheng X., Mallette F.A., Watowich S.S., and Sun S.C.. 2014. Noncanonical NF-κB pathway controls the production of type I interferons in antiviral innate immunity. Immunity. 40:342–354. 10.1016/j.immuni.2014.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, P.A., Ohtani H., Chakravarthy A., and De Carvalho D.D.. 2019. Epigenetic therapy in immune-oncology. Nat. Rev. Cancer. 19:151–161. 10.1038/s41568-019-0109-9 [DOI] [PubMed] [Google Scholar]
- Jung, H., Kim H.S., Kim J.Y., Sun J.-M., Ahn J.S., Ahn M.-J., Park K., Esteller M., Lee S.-H., and Choi J.K.. 2019. DNA methylation loss promotes immune evasion of tumours with high mutation and copy number load. Nat. Commun. 10:4278. 10.1038/s41467-019-12159-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, X., Chen J., Xie W., Brown S.M., Cai Y., Wu K., Fan D., Nie Y., Yegnasubramanian S., Tiedemann R.L., et al. 2019. Defining UHRF1 Domains that Support Maintenance of Human Colon Cancer DNA Methylation and Oncogenic Properties. Cancer Cell. 35:633–648.e7. 10.1016/j.ccell.2019.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladle, B.H., Li K.-P., Phillips M.J., Pucsek A.B., Haile A., Powell J.D., Jaffee E.M., Hildeman D.A., and Gamper C.J.. 2016. De novo DNA methylation by DNA methyltransferase 3a controls early effector CD8+ T-cell fate decisions following activation. Proc. Natl. Acad. Sci. USA. 113:10631–10636. 10.1073/pnas.1524490113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, A.J., and Ashkar A.A.. 2018. The Dual Nature of Type I and Type II Interferons. Front. Immunol. 9:2061. 10.3389/fimmu.2018.02061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, P.P., Fitzpatrick D.R., Beard C., Jessup H.K., Lehar S., Makar K.W., Pérez-Melgosa M., Sweetser M.T., Schlissel M.S., Nguyen S., et al. 2001. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 15:763–774. 10.1016/S1074-7613(01)00227-8 [DOI] [PubMed] [Google Scholar]
- Leung, Y.T., Shi L., Maurer K., Song L., Zhang Z., Petri M., and Sullivan K.E.. 2015. Interferon regulatory factor 1 and histone H4 acetylation in systemic lupus erythematosus. Epigenetics. 10:191–199. 10.1080/15592294.2015.1009764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyko, F. 2018. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 19:81–92. 10.1038/nrg.2017.80 [DOI] [PubMed] [Google Scholar]
- Maggirwar, S.B., Harhaj E.W., and Sun S.C.. 1997. Regulation of the interleukin-2 CD28-responsive element by NF-ATp and various NF-kappaB/Rel transcription factors. Mol. Cell. Biol. 17:2605–2614. 10.1128/MCB.17.5.2605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen, S.S., Jensen S.B., Chiliveru S., Melchjorsen J., Julkunen I., Gaestel M., Arthur J.S.C., Flavell R.A., Ghosh S., and Paludan S.R.. 2009. RIG-I-mediated activation of p38 MAPK is essential for viral induction of interferon and activation of dendritic cells: dependence on TRAF2 and TAK1. J. Biol. Chem. 284:10774–10782. 10.1074/jbc.M807272200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Nebreda, L., McLafferty F.S., and Singer B.D.. 2019. DNA methylation as a transcriptional regulator of the immune system. Transl. Res. 204:1–18. 10.1016/j.trsl.2018.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee, S., Vipat V.C., and Chakrabarti A.K.. 2013. Infection with influenza A viruses causes changes in promoter DNA methylation of inflammatory genes. Influenza Other Respir. Viruses. 7:979–986. 10.1111/irv.12127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsukasa, H., Oda M., Yin J., Chikuma S., Ito M., Koga-Iizuka M., Someya K., Kitagawa Y., Ohkura N., Sakaguchi S., et al. 2019. Loss of TET proteins in regulatory T cells promotes abnormal proliferation, Foxp3 destabilization and IL-17 expression. Int. Immunol. 31:335–347. 10.1093/intimm/dxz008 [DOI] [PubMed] [Google Scholar]
- National Genomics Data Center Members and Partners . 2020. Database Resources of the National Genomics Data Center in 2020. Nucleic Acids Res. 48(D1):D24–D33. 10.1093/nar/gkz913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, J.D., Denisenko O., and Bomsztyk K.. 2006. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat. Protoc. 1:179–185. 10.1038/nprot.2006.27 [DOI] [PubMed] [Google Scholar]
- Niewold, T.B. 2014. Type I interferon in human autoimmunity. Front. Immunol. 5:306. 10.3389/fimmu.2014.00306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obata, Y., Furusawa Y., Endo T.A., Sharif J., Takahashi D., Atarashi K., Nakayama M., Onawa S., Fujimura Y., Takahashi M., et al. 2014. The epigenetic regulator Uhrf1 facilitates the proliferation and maturation of colonic regulatory T cells. Nat. Immunol. 15:571–579. 10.1038/ni.2886 [DOI] [PubMed] [Google Scholar]
- Okano, M., Bell D.W., Haber D.A., and Li E.. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 99:247–257. 10.1016/S0092-8674(00)81656-6 [DOI] [PubMed] [Google Scholar]
- Pacis, A., Tailleux L., Morin A.M., Lambourne J., MacIsaac J.L., Yotova V., Dumaine A., Danckaert A., Luca F., Grenier J.-C., et al. 2015. Bacterial infection remodels the DNA methylation landscape of human dendritic cells. Genome Res. 25:1801–1811. 10.1101/gr.192005.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paludan, S.R., and Bowie A.G.. 2013. Immune sensing of DNA. Immunity. 38:870–880. 10.1016/j.immuni.2013.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry, A.K., Chen G., Zheng D., Tang H., and Cheng G.. 2005. The host type I interferon response to viral and bacterial infections. Cell Res. 15:407–422. 10.1038/sj.cr.7290309 [DOI] [PubMed] [Google Scholar]
- Qi, S., Li Y., Dai Z., Xiang M., Wang G., Wang L., and Wang Z.. 2019. Uhrf1-Mediated Tnf-α Gene Methylation Controls Proinflammatory Macrophages in Experimental Colitis Resembling Inflammatory Bowel Disease. J. Immunol. 203:3045–3053. 10.4049/jimmunol.1900467 [DOI] [PubMed] [Google Scholar]
- Rasmussen, K.D., and Helin K.. 2016. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 30:733–750. 10.1101/gad.276568.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin, A., and Cedar H.. 1991. DNA methylation and gene expression. Microbiol. Rev. 55:451–458. 10.1128/MR.55.3.451-458.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samata, M., Alexiadis A., Richard G., Georgiev P., Nuebler J., Kulkarni T., Renschler G., Basilicata M.F., Zenk F.L., Shvedunova M., et al. 2020. Intergenerationally Maintained Histone H4 Lysine 16 Acetylation Is Instructive for Future Gene Activation. Cell. 182:127–144.e23. 10.1016/j.cell.2020.05.026 [DOI] [PubMed] [Google Scholar]
- Schlosberg, C.E., VanderKraats N.D., and Edwards J.R.. 2017. Modeling complex patterns of differential DNA methylation that associate with gene expression changes. Nucleic Acids Res. 45:5100–5111. 10.1093/nar/gkx078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selinger, E., and Reiniš M.. 2018. Epigenetic View on Interferon γ Signalling in Tumour Cells. Folia Biol. (Praha). 64:125–136. [PubMed] [Google Scholar]
- Shestakova, E.A. 2015. Different mechanisms of epigenetic regulation of gene expression. MOJ Cell Sci. Rep. 2:10–14. 10.15406/mojcsr.2015.02.00019 [DOI] [Google Scholar]
- Stetson, D.B., and Medzhitov R.. 2006. Type I interferons in host defense. Immunity. 25:373–381. 10.1016/j.immuni.2006.08.007 [DOI] [PubMed] [Google Scholar]
- Suarez-Alvarez, B., Rodriguez R.M., Fraga M.F., and López-Larrea C.. 2012. DNA methylation: a promising landscape for immune system-related diseases. Trends Genet. 28:506–514. 10.1016/j.tig.2012.06.005 [DOI] [PubMed] [Google Scholar]
- Sun, F., Abreu-Rodriguez I., Ye S., Gay S., Distler O., Neidhart M., and Karouzakis E.. 2019. TET1 is an important transcriptional activator of TNFα expression in macrophages. PLoS One. 14:e0218551. 10.1371/journal.pone.0218551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi, O., and Akira S.. 2010. Pattern recognition receptors and inflammation. Cell. 140:805–820. 10.1016/j.cell.2010.01.022 [DOI] [PubMed] [Google Scholar]
- Tang, R.-Z., Zhu J.-J., Yang F.-F., Zhang Y.-P., Xie S.-A., Liu Y.-F., Yao W.-J., Pang W., Han L.-L., Kong W., et al. 2019. DNA methyltransferase 1 and Krüppel-like factor 4 axis regulates macrophage inflammation and atherosclerosis. J. Mol. Cell. Cardiol. 128:11–24. 10.1016/j.yjmcc.2019.01.009 [DOI] [PubMed] [Google Scholar]
- Thomas, R.M., Gamper C.J., Ladle B.H., Powell J.D., and Wells A.D.. 2012. De novo DNA methylation is required to restrict T helper lineage plasticity. J. Biol. Chem. 287:22900–22909. 10.1074/jbc.M111.312785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topper, M.J., Vaz M., Chiappinelli K.B., DeStefano Shields C.E., Niknafs N., Yen R.C., Wenzel A., Hicks J., Ballew M., Stone M., et al. 2017. Epigenetic Therapy Ties MYC Depletion to Reversing Immune Evasion and Treating Lung Cancer. Cell. 171:1284–1300.e21. 10.1016/j.cell.2017.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsagaratou, A., González-Avalos E., Rautio S., Scott-Browne J.P., Togher S., Pastor W.A., Rothenberg E.V., Chavez L., Lähdesmäki H., and Rao A.. 2017. TET proteins regulate the lineage specification and TCR-mediated expansion of iNKT cells. Nat. Immunol. 18:45–53. 10.1038/ni.3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urdinguio, R.G., Lopez V., Bayón G.F., Diaz de la Guardia R., Sierra M.I., García-Toraño E., Perez R.F., García M.G., Carella A., Pruneda P.C., et al. 2019. Chromatin regulation by Histone H4 acetylation at Lysine 16 during cell death and differentiation in the myeloid compartment. Nucleic Acids Res. 47:5016–5037. 10.1093/nar/gkz195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Hu, H., and Zhang, K.. 2017. Overview of Interferon: Characteristics, signaling and anti-cancer effect. Arch. Biotechnol. Biomed. 1:001–016. 10.29328/journal.hjb.1001001 [DOI] [Google Scholar]
- Wang, J., Basagoudanavar S.H., Wang X., Hopewell E., Albrecht R., García-Sastre A., Balachandran S., and Beg A.A.. 2010. NF-kappa B RelA subunit is crucial for early IFN-beta expression and resistance to RNA virus replication. J. Immunol. 185:1720–1729. 10.4049/jimmunol.1000114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L., Liu Y., Beier U.H., Han R., Bhatti T.R., Akimova T., and Hancock W.W.. 2013. Foxp3+ T-regulatory cells require DNA methyltransferase 1 expression to prevent development of lethal autoimmunity. Blood. 121:3631–3639. 10.1182/blood-2012-08-451765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., Cao Q., Yu L., Shi H., Xue B., and Shi H.. 2016. Epigenetic regulation of macrophage polarization and inflammation by DNA methylation in obesity. JCI Insight. 1:e87748–e87748. 10.1172/jci.insight.87748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Song F., Zhu J., Zhang S., Yang Y., Chen T., Tang B., Dong L., Ding N., Zhang Q., et al. 2017. GSA: Genome Sequence Archive. Genomics Proteomics Bioinformatics. 15(1):14–18. 10.1016/j.gpb.2017.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, E., and Wei C.-L.. 2011. Genome-wide distribution of dna methylation at single-nucleotide resolution. In Progress in Molecular Biology and Translational Science. Cheng X., and Blumenthal R.M., editors. Academic Press, Cambridge. 459–477. 10.1016/B978-0-12-387685-0.00015-9 [DOI] [PubMed] [Google Scholar]
- Yang, X., Wang X., Liu D., Yu L., Xue B., and Shi H.. 2014. Epigenetic regulation of macrophage polarization by DNA methyltransferase 3b. Mol. Endocrinol. 28:565–574. 10.1210/me.2013-1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, R., Qu C., Zhou Y., Konkel J.E., Shi S., Liu Y., Chen C., Liu S., Liu D., Chen Y., et al. 2015. Hydrogen Sulfide Promotes Tet1- and Tet2-Mediated Foxp3 Demethylation to Drive Regulatory T Cell Differentiation and Maintain Immune Homeostasis. Immunity. 43:251–263. 10.1016/j.immuni.2015.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Q., Zhou B., Zhang Y., Nguyen E.T., Du J., Glosson N.L., and Kaplan M.H.. 2012. DNA methyltransferase 3a limits the expression of interleukin-13 in T helper 2 cells and allergic airway inflammation. Proc. Natl. Acad. Sci. USA. 109:541–546. 10.1073/pnas.1103803109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., Qiu Y., Yang J., Bian S., Chen G., Deng M., Kang H., and Huang L.. 2016. DNMT1-PPARγ pathway in macrophages regulates chronic inflammation and atherosclerosis development in mice. Sci. Rep. 6:30053–30053. 10.1038/srep30053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue, X., Lio C.J., Samaniego-Castruita D., Li X., and Rao A.. 2019. Loss of TET2 and TET3 in regulatory T cells unleashes effector function. Nat. Commun. 10:2011. 10.1038/s41467-019-09541-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q., Zhao K., Shen Q., Han Y., Gu Y., Li X., Zhao D., Liu Y., Wang C., Zhang X., et al. 2015. Tet2 is required to resolve inflammation by recruiting Hdac2 to specifically repress IL-6. Nature. 525:389–393. 10.1038/nature15252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, T., Zhang Z., Dong Q., Xiong J., and Zhu B.. 2020. Histone H3K27 acetylation is dispensable for enhancer activity in mouse embryonic stem cells. Genome Biol. 21:45. 10.1186/s13059-020-01957-w [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists DEGs between WT and Uhrf1-deficient BMDMs without stimulation.
lists the DEGs between WT and Uhrf1-deficient BMDMs stimulated with 20 µg/ml pI:C for 2 h assessed by mRNA sequencing.
lists ATAC-seq of WT and Uhrf1-deficient BMDMs without stimulation.
lists ATAC-seq of WT and Uhrf1-deficient BMDMs under pI:C stimulation.
lists the sequences of genotyping primers.
lists the sequences of probes used for EMSA.
lists the oligonucleotide sequences used for qPCR.
Data Availability Statement
The raw sequence data reported in this paper were deposited in the Genome Sequence Archive (Wang et al., 2017) at the BIG Data Center (National Genomics Data Center Members and Partners, 2020), Beijing Institute of Genomics, Chinese Academy of Sciences, under the accession no. PRJCA002630.