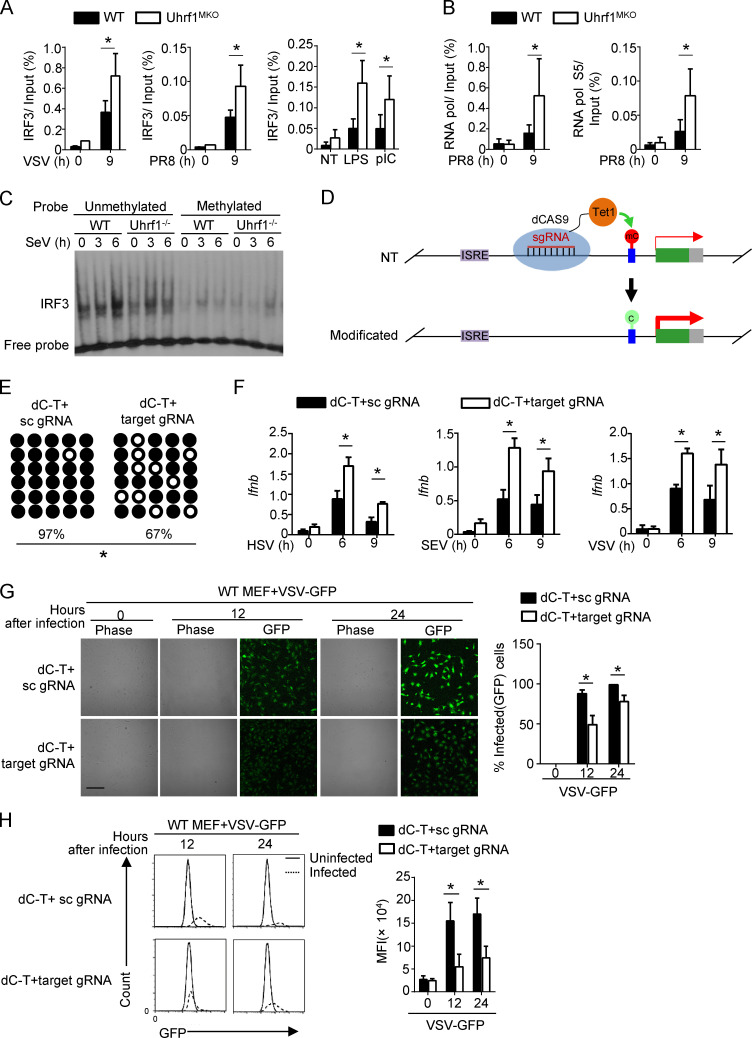
Figure 6.
Targeting DNA methylation editing by dCas9-Tet1 activates IFN-I production. WT and Uhrf1-deficient BMDMs were stimulated for 9 h with specific virus or TLR agonists. (A and B) ChIP assays were performed and quantified by qPCR to detect the binding of IRF3 (A) or RNA polymerase (RNA pol; B) to the Ifnb promoter (n = 4). (C) EMSA of nuclear extracts of WT and Uhrf1-deficient BMDMs stimulated with Sendai Virus, as assessed using an unmethylated or methylated HRP-labeled Ifnb promoter probe. (D) Schematic graph of a catalytically inactive mutant Cas9 (dCas9) fused to Tet1 to remove DNA methylation modifications from the Ifnb promoter. (E) Bisulfite sequencing of WT MEFs transfected with dCas9-Tet1 (dC-T) plus a scrambled gRNA (sc gRNA) or four gRNAs targeting the Ifnb promoter region (target gRNA). (F) The mRNA levels of Ifnb induced by different viruses in MEFs described in E were measured by qPCR (n = 3). (G and H) MEFs described in B were infected with VSV-GFP at an MOI of 0.1 for 24 h. The data are presented as a representative picture, showing the infected (GFP+) and total (bright-field) cells (G; n = 3). Scale bar, 1,000 µm. Summary graph of flow cytometric quantification of the infected cells (H; n = 4). The data in the qPCR assay are presented as fold changes relative to the actin mRNA levels. All data are representative of at least three independent experiments. The data are presented as means ± SEMs. The significance of differences was determined by a t test. *, P < 0.05.