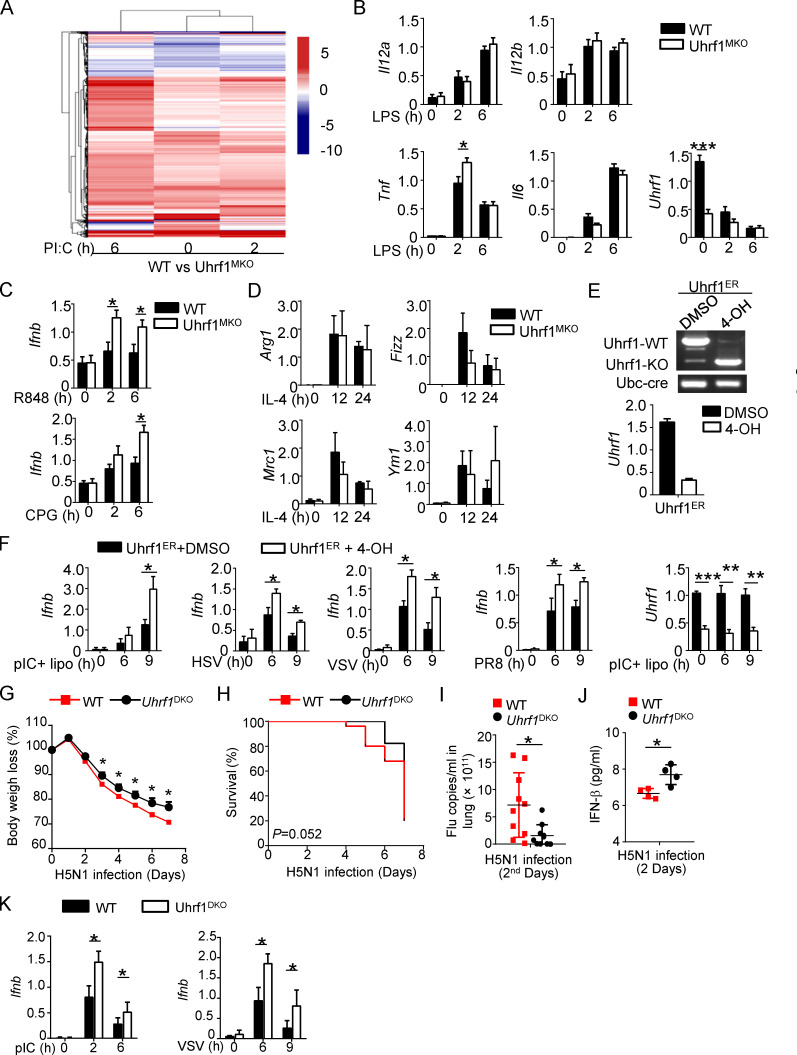
Figure S3.
Uhrf1 is not required for the proinflammatory cytokines production except Ifnb. (A) Heatmap showing the pI:C-stimulated DEGs in BMDMs from WT and Uhrf1MKO mice. The DEGs were identified with a fold change of experimental sample to nontreated control (ES/NT) >2.0 or <0.5. (B) qRT-PCR analysis of the indicated genes using WT or Uhrf1-deficient BMDMs stimulated with 100 ng/ml LPS (n = 6). (C) Ifnb mRNA in Uhrf1MKO BMDMs were measured by qPCR assay responding to 1 μg/ml R848 or 5 μM CpG (n = 4). (D) qRT-PCR analysis of the indicated genes using WT or Uhrf1-deficicent BMDMs stimulated with 10 ng/ml IL-4 (n = 3). (E) Uhrf1ER-Cre MEFs were incubated with DMSO or 4-OH for 72 h, and we examined the genotyping by amplifying WT and KO alleles. qRT-PCR analysis of the Uhrf1 mRNA level. (F) qRT-PCR analysis of the indicated genes using WT or Uhrf1-deficient MEFs generated as above stimulated with pI:C plus 4-OH (n = 4). (G–J) WT and DC-conditional Uhrf1 KO (Uhrf1DKO) mice (6–8 wk) were i.n. infected with a sublethal dose (0.1 HA) of H5N1 influenza virus. Body weight loss (G) and survival rate (H) were measured for 14 d (n = 17). (I) Viral titers in the lung were quantified on day 2 by a TCID50 assay (n = 10). (J) ELISA for IFN-β in the sera of WT and Uhrf1MKO mice infected with H5N1 influenza virus on days 2 and 5 (n = 4). (K) qRT-PCR analysis of the indicated genes using WT or Uhrf1-deficient BM-derived DCs stimulated with 100 ng/ml LPS or 1 MOI VSV. Data in the qPCR assay are presented as fold relative to the actin mRNA level. All data are representative of at least three independent experiments. Data are presented as means ± SEMs. The significances of differences were determined by a t test. *, P < 0.05; **, P < 0.01; ***, P < 0.005.