Figure 3.
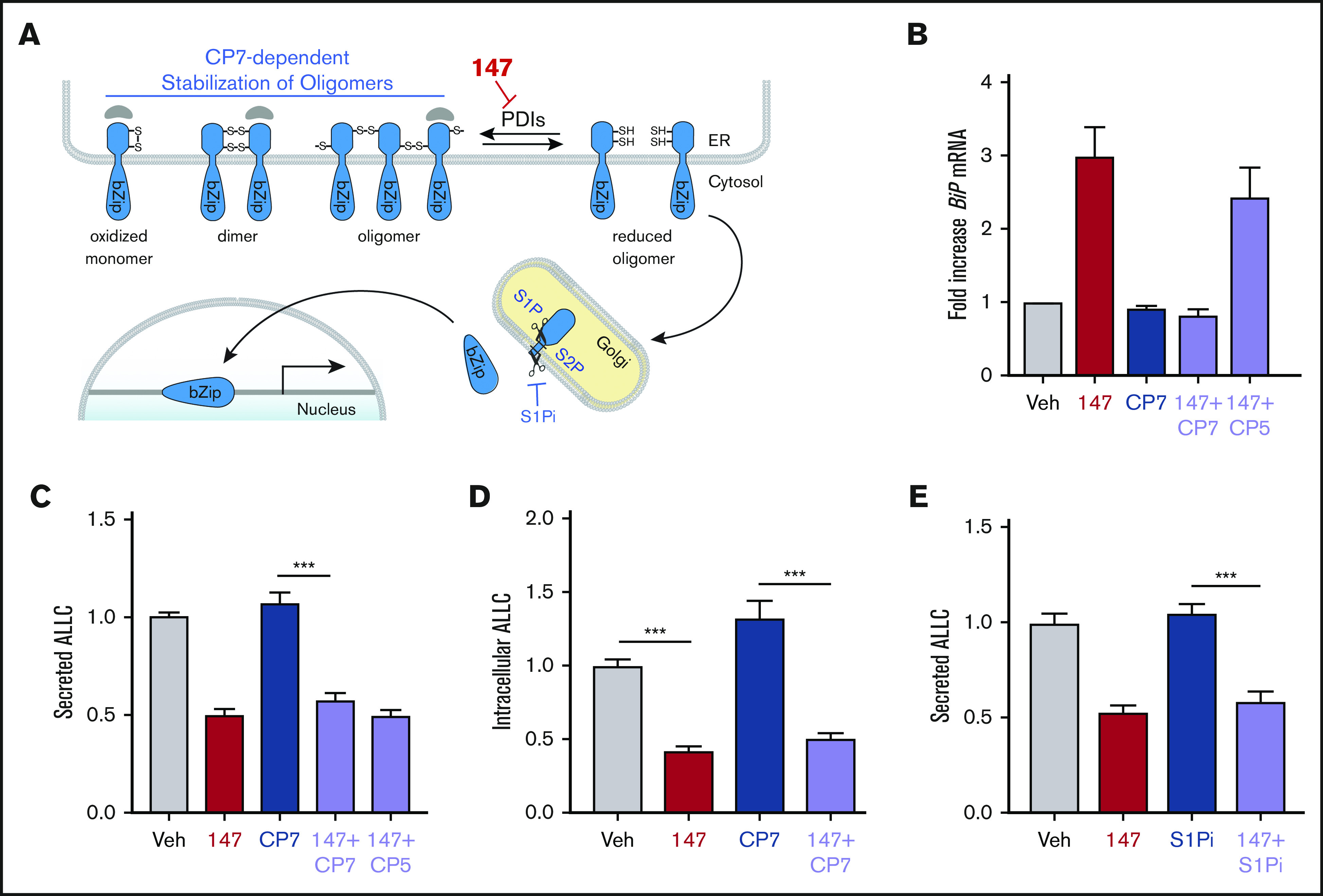
147-dependent reductions in ALLC secretion are independent of ATF6 activation. (A) Illustration showing the mechanism of 147-dependent ATF6 activation. ATF6 is retained within the ER as oxidized monomers, disulfide-bonded dimers, or disulfide-bonded oligomers that are maintained by PDIs. Compound 147 covalently modifies a subset of PDIs, increasing the population of reduced ATF6 monomers that can traffic to the Golgi and undergo proteolytic processing by S1P and S2P. This releases the N-terminal bZIP domain of ATF6 to localize to the nucleus and promote ATF6 transcriptional signaling. The specific steps of this activation mechanism inhibited by CP7 or site 1 protease (S1P) inhibitor (S1Pi) are shown. (B) Graph showing BiP expression measured by quantitative polymerase chain reaction in ALMC-2 cells treated for 5 hours with vehicle (Veh) or 147 (10 µM), CP7 (10 µM), or CP5 (10 µM), as indicated. Error bars show 95% confidence interval for 3 replicates. (C) Bar graphs showing normalized amounts of ALLC in conditioned media prepared from ALMC-2 cells treated for 18 hours with Veh or 147 (10 µM), CP7 (10 µM), or CP5 (10 µM), as indicated. ALLC was quantified by ELISA. Error bars show standard error of the mean (SEM) for 21 replicates across 4 independent experiments. (D) Bar graphs showing normalized amounts of ALLC in lysates prepared from ALMC-2 cells treated for 18 hours with Veh, 147 (10 µM), and/or CP7 (10 µM), as indicated. ALLC was quantified by ELISA. Error bars show SEM for 10 replicates across 2 independent experiments. (E) Bar graphs showing normalized amounts of ALLC in conditioned media from ALMC-2 cells treated for 18 hours with Veh or 147 (10 µM) and/or S1Pi PF429242 (10 µM), as indicated. ALLC was quantified by ELISA. Error bars show SEM for 8 replicates across 2 independent experiments. ***P < .005 (unpaired Student t test). mRNA, messenger RNA.
