Figure 2. Sequencing and Analytical Approaches to Identify Genes Associated With Exfoliation Syndrome.
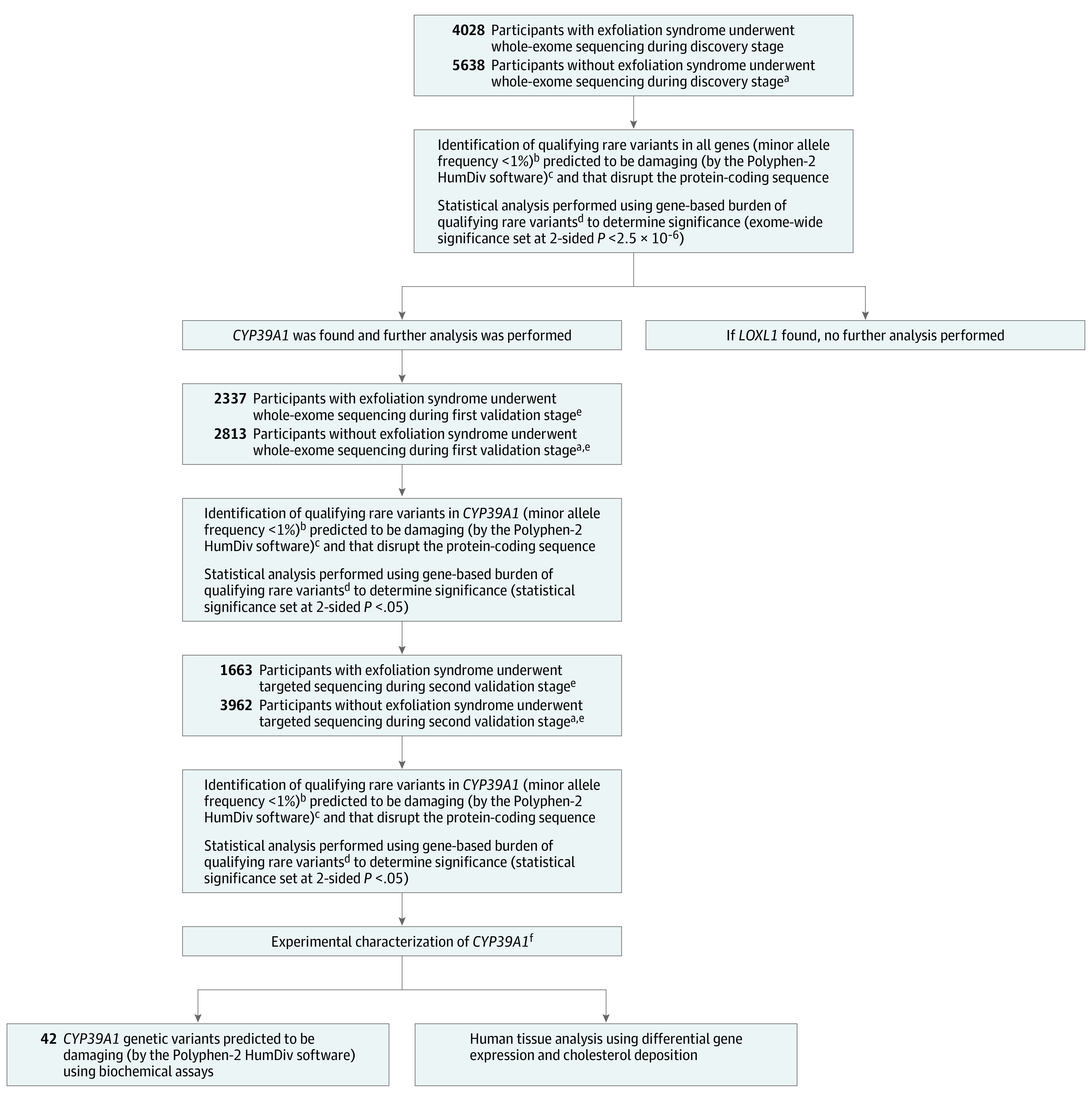
aEach participant with exfoliation syndrome was matched to at least 1 participant without exfoliation syndrome (for every individual with exfoliation syndrome, ≥1 individual without exfoliation syndrome could be recruited as a matching control). Matching was by geographical site of recruitment and self-reported ancestry.
bMost disease-causing genetic variants are maintained at low frequencies by purifying evolutionary selection.
cPolymorphism Phenotyping version 2 (Polyphen-2) is a widely used computer algorithm to identify genetic variants that damage protein function. Such variants are likely to cause disease. Alleles predicted to be benign were excluded because their inclusion could have masked disease associations caused by damaging allelic variants.
dMany medical conditions are caused by haploinsufficiency, which is a state where 1 copy of a gene has been damaged by alterations, leaving the remaining normally functioning gene copy insufficient to sustain normal function. In many of these conditions, different alterations within a given gene causes the same damaging consequences to the encoded protein product. In this study, the burden test was designed to compare the number (or burden) of damaging genetic alterations found in each gene among persons with exfoliation syndrome vs those without exfoliation syndrome.
eValidation of original discovery stage findings in 2 independently enrolled validation cohorts increases the confidence that the original observations were not false discoveries.
fGenes identified to be significantly associated with disease during the discovery and validation stages were characterized further using post hoc experimental assays to garner additional insights into disease pathogenesis.
