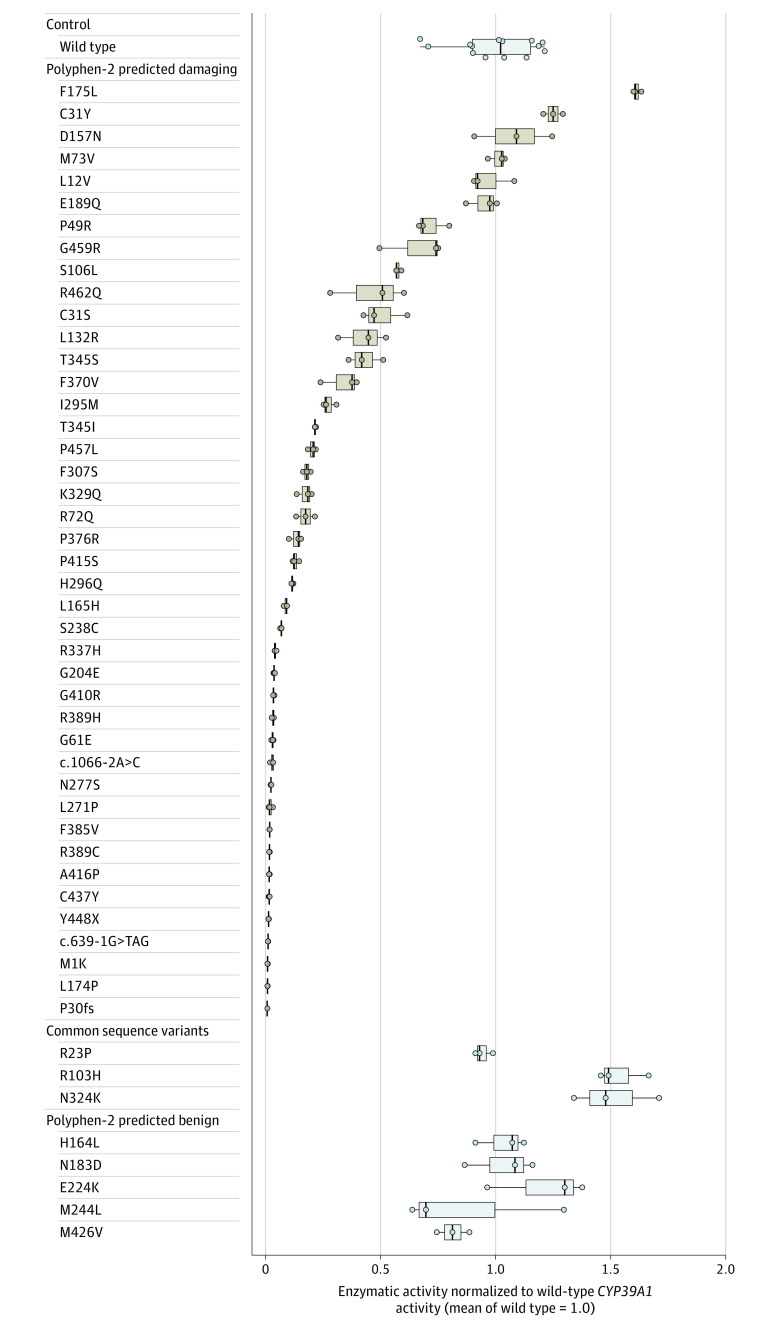
Figure 4. Experimental Determination of Functional Enzymatic Activity for CYP39A1 Variants.
Box and whisker plots of functional enzymatic activity of 1 wild-type CYP39A1 variant, 3 common variants, 42 rare variants predicted to be damaging (by Polymorphism Phenotyping version 2 [Polyphen-2]; software that predicts the possible effect of amino acid substitutions on human protein function), and 5 variants predicted by Polyphen-2 to be benign. Three data points (representing 3 independent biological replicates) were plotted for all tested CYP39A1 genetic variants. The crossbars represent the median level of activity, the boxes represent the interquartile range, and the whiskers portray the range of enzymatic activity for each variant. Individual biological replicates are shown as individual data points. Amino acid substitutions are caused by coding-sequence genetic variation. The case-control distribution of all variants predicted by Polyphen-2 to be damaging appears in eTable 10 in Supplement 1. The enzymatic activity of individual variants was expressed as a proportion relative to the wild-type CYP39A1 enzymatic activity. The functional enzymatic activity of each tested CYP39A1 variant was determined by the relative 24(S)-7α,24-dihydroxycholesterol product abundance it produced with respect to wild-type CYP39A1. The product abundance was calculated by integrating the area under the 24(S)-7α,24-dihydroxycholesterol product peak (eFigure 10 in Supplement 1), averaged across 3 independent samples for each tested variant. The empirical distribution of the wild-type CYP39A1 enzymatic activity was defined using 14 independent biological replicates. The design and validation of the experimental system appear in eFigures 9 and 10 in Supplement 1.