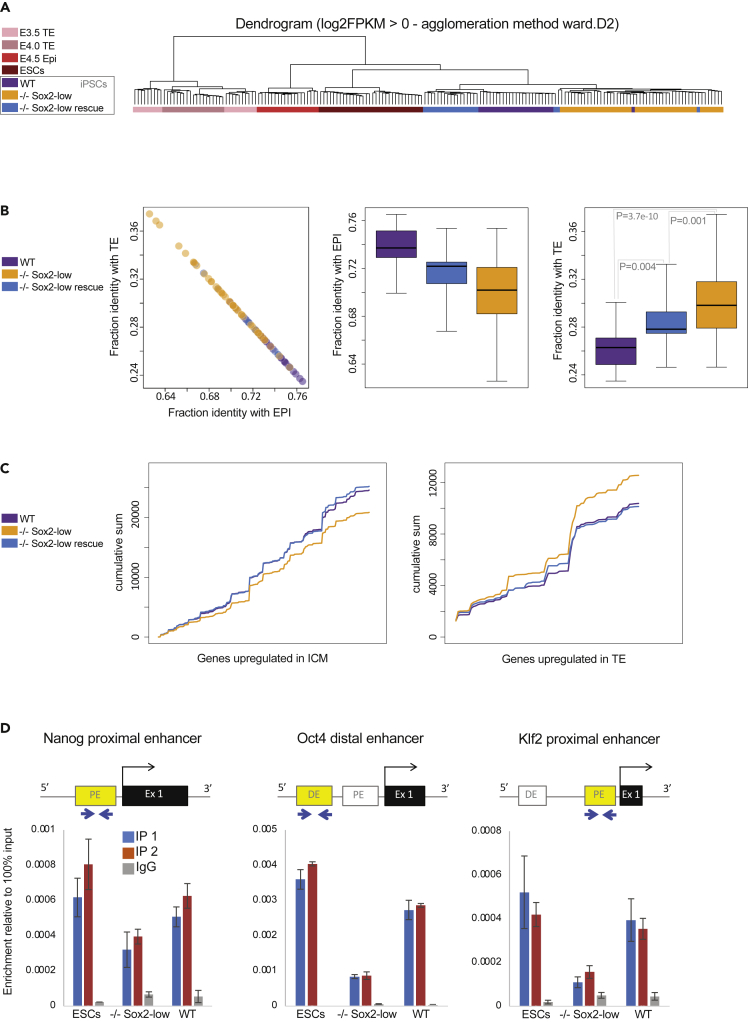
Figure 6.
Sox2-low iPSCs have weaker epiblast identity
(A) Dendrogram computed with the top variable genes among the selected cell types (FPKM >1, logCV2 > 0.5, n = 1,446). Trophectoderm/trophoblast (TE) E3.5 and E4.0 Single-cell RNA-sequencing (scRNA-seq) data is from (Deng et al., 2014), E4.5 Epiblast (Epi) scRNA-seq data is from (Mohammed et al., 2017), scRNA-seq ESC data is from (Sousa et al., 2018).
(B) Scatterplot of fraction of identity between WT, −/− Sox2-low, −/− Sox2-low rescue iPSC line samples and embryo lineages (Epi and TE). Boxplot of the distribution of the fraction of identity between each population. Student's t-test was applied to calculate the significance of the differences in similarity to TE identity between −/−Sox2-low, WT and −/−Sox2-low rescue iPSCs.
(C) Cumulative sum for genes upregulated in TE or ICM (Blakeley et al., 2015), computed with gene expression value for WT, −/− Sox2-low, −/− Sox2-low rescue iPSC line samples.
(D) Oct4 chromatin immunoprecipitation in Rex1-GFP ESCs and iPSCs in 2iLIF. IP = immunoprecipitation; IgG = negative control. Error bars represent the standard deviation of three technical qPCR replicates. IP1 and IP2 represent independent Oct4 immunoprecipitations. IgG represents negative control normal IgG immunoprecipitation.