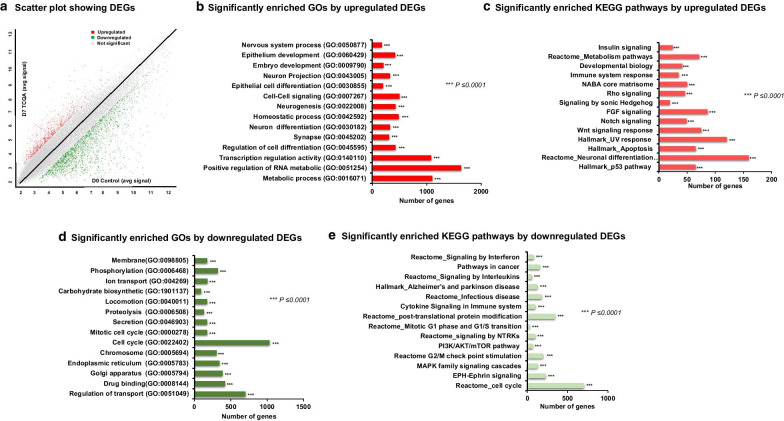
Fig. 1.
Microarray profiling of D7 TCQA-treated versus D0 control hAECs. a Scatter plot showing the DEGs. X-axis represents the average signal intensities (log2) in D0 control. Y-axis represents the average signal intensities (log2) in D7 TCQA-treated hAECs. The red color represents the upregulated DEGs, green color represents the downregulated DEGs, and grey color represents the nonsignificant genes. b Significantly enriched GOs by upregulated (analyzed using GSEA). c Significantly enriched KEGG pathways by upregulated DEGs (analyzed using DAVID and GSEA). d Significantly enriched GOs by downregulated (analyzed using GSEA). e Top significantly enriched KEGG pathways by the downregulated DEGs (analyzed using DAVID and GSEA). Each bar is arranged according to significance (p-values) and represents the number of DEGs