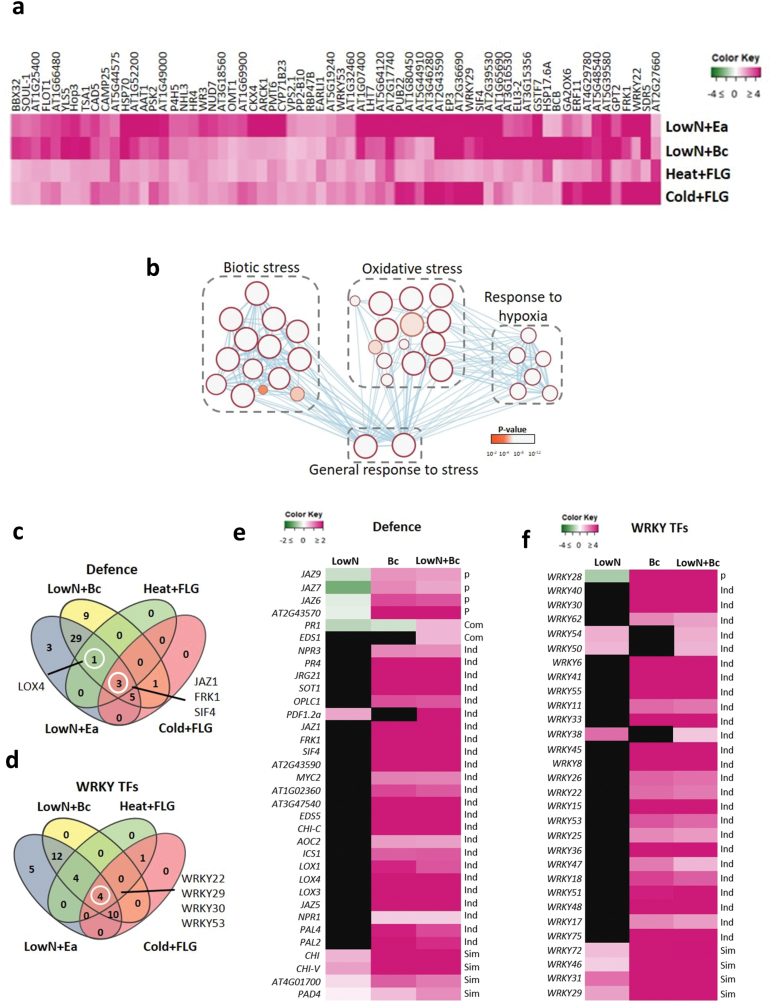
Fig. 3.
Meta-analysis of combined abiotic and biotic stress transcriptome data. Ten publicly available datasets were selected to study the modulation of defense gene expression in response to combined stresses. Transcriptome data for nitrate limitation (LowN), Botrytis cinerea (Bc), and their combination were obtained from Soulié et al. (2020), whilst the data for cold and flagellin (FLG) and for heat and FLG were extracted from the Gene Expression Omnibus repository (accession GSE41935; Rasmussen et al., 2013). (a) Expression of 66 commonly up-regulated genes in all four multistress conditions (fold change >1; see Supplementary Table S2). (b) Gene Ontology analysis was performed using the Cytoscape and g:Profiler software according to Reimand et al. (2019) using the total of 197 up- and down-regulated genes shared in all multistress conditions (fold-change >1 or <–1; Supplementary Table S2). (c. d) Venn diagrams showing (c) three defense genes and (d) four WRKYs commonly modulated by all the combined stress conditions. (e, f) Heat-maps showing expression of selected defense genes (e) and WRKY transcription factors (d), following single LowN stress, Bc infection, and their combinations. Black shading indicates genes not significantly modulated (P>0.05; Supplementary Tables S2, S3). The same genes reported in the heat-maps were also screened in datasets related to the combination of heat, cold, and flagellin (see Supplementary Table S3).