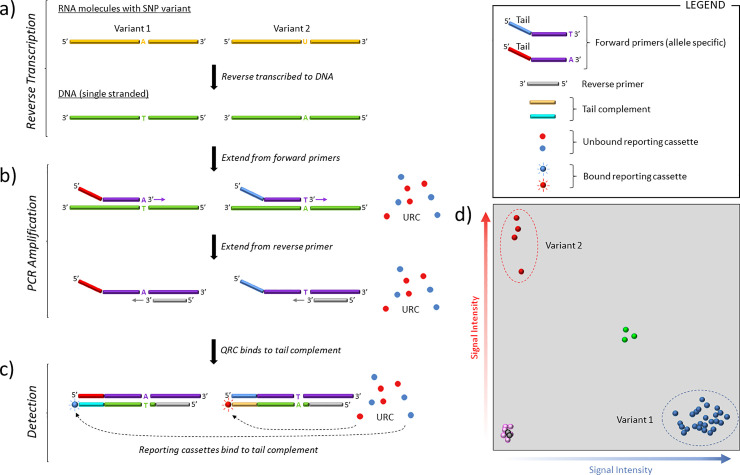
Fig 2. PACE-RT™ genotyping.
a) The RNA sample, which here contains two SNP variants, is reverse transcribed to produce single stranded DNA. b) The target region is then amplified using two, allele specific primers which differ in their 3’ terminal base and 5’ tail, and a common reverse primer. As PCR proceeds, the tail sequences of allele-specific forward primers become incorporated in the amplified fragments and their sequence complements are generated. c) Reporting cassettes, initially quenched, bind to the appropriate tail sequence complement, become unquenched and produce a light signal (HEX (red), FAM (blue)). d) Fluorescence intensity is measured and plotted to determine allele calls. A mixed signal (green) is seen as equal signal from both fluorochromes. Samples at the origin are the no-template controls (black) and unamplified samples (pink).