Fig. 5. LINC01569 mediates mechanosensor mRNA decay via GMD manner.
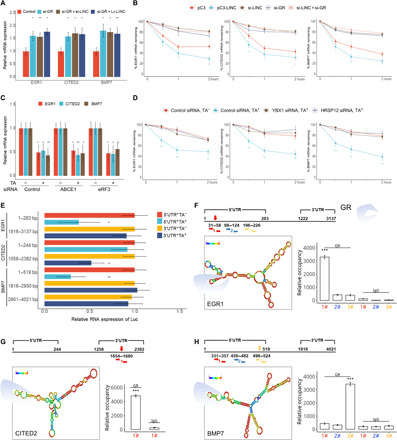
(A) Stretched HSFBs were treated with TA and transfected with control siRNAs or those targeting the indicated genes, followed by qRT-PCR. (B) Stretched HSFBs were transfected with a control (pC3) or LINC01569-overexpressing vector (pC3-LINC) and/or with the indicated siRNAs. Then, the cells were treated with actinomycin D (ACD) (5 μg/ml), and total cellular RNAs were extracted at the indicated time points for qRT-PCR to measure the half-lives of endogenous mechanosensors. The normalized level of each mechanosensor measured at 0 hours was set as 100%. The y axis represents a logarithmic scale of the levels of remaining mRNA (in percentage). (C) Stretched HSFBs were treated with or without TA and transfected with the indicated siRNAs, followed by qRT-PCR assay. (D) Stretched HSFBs were transfected with si-YBX1 or si-HRSP12 with or without TA treatment. The levels of remaining mRNA were analyzed as described in (B). (E) HSFBs were transfected with the luciferase (Luc) reporter constructs for the indicated mRNA regions of the mechanosensors. Cells were treated with or without TA and subjected to qRT-PCR analysis for luciferase mRNA levels. Data are normalized to GAPDH mRNA expression. (F to H) Top: The predicted RNA secondary structure of the hairpin formation in the indicated mRNAs. Red arrows indicate potential GR-binding regions. Bottom: The enrichment of GR-binding sequences on the indicated mRNAs, as assessed using PAR-CLIP (photoactivatable ribonucleoside–enhanced cross-linking and immunoprecipitation) quantitative PCR (qPCR). For (A) to (H), data are presented as means ± SD. *P < 0.05, **P < 0.01, and ***P < 0.001, by ANOVA for more than two groups.
