Fig. 7. LINC01569 recruits YBX1 to target mRNAs in the GMD complex.
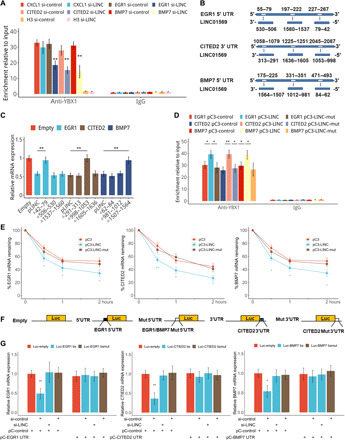
(A) Stretched HSFBs were treated with TA and transfected with the control or LINC01569 siRNAs. The cells were used in an IP experiment performed using an anti-YBX1 antibody (rabbit IgG as a negative control). EGR1, CITED2, and BMP7 mRNA levels were evaluated. (B) Schematic representation of the predicted binding sites of LINC01569 on the UTRs of EGR1, CITED2, and BMP7. (C) TA-treated and stretched HSFBs were transfected with empty vectors or those expressing wild-type or truncated LINC01569 and were further treated with ACD. The levels of indicated mRNAs were measured using qRT-PCR. (D) TA-treated and stretched HSFBs were transfected with the constructs for wild-type LINC01569 or LINC01569 harboring mutations (mut) in the sites responsible for binding to the corresponding mRNAs. RIP was performed using an anti-YBX1 antibody. The immunoprecipitated RNA was extracted and subjected to reverse transcription. (E) HSFBs were transfected with empty vectors or those overexpressing wild-type LINC01569 or LINC01569 harboring mutations in the mRNA binding sites, followed by treatment with ACD. The mRNAs of mechanosensors were measured using qRT-PCR. (F) Schematic representation of the UTR regions of the mechanosensors cloned into the luciferase plasmid. (G) Luciferase vectors containing wild-type or mutated LINC01569-binding sequences of EGR1, CITED2, and BMP7 UTRs and siRNAs were introduced into TA-treated and stretched HSFBs. pCDNA expressing EGR1, CITED2, and BMP7 UTRs were cotransfected as competitors on binding to LINC01569 with luciferase transcripts containing EGR1, CITED2, and BMP7 UTRs. Luciferase mRNA expression was determined by qPCR 48 hours after transfection. Data were normalized to GAPDH mRNA expression. For (A), (C), (D), (E), and (G), data are presented as means ± SD. *P < 0.05 and **P < 0.01, by Student’s t test for two groups or ANOVA for more than two groups.
