Figure 4. Dynamic Interactions of NusG in the PTC.
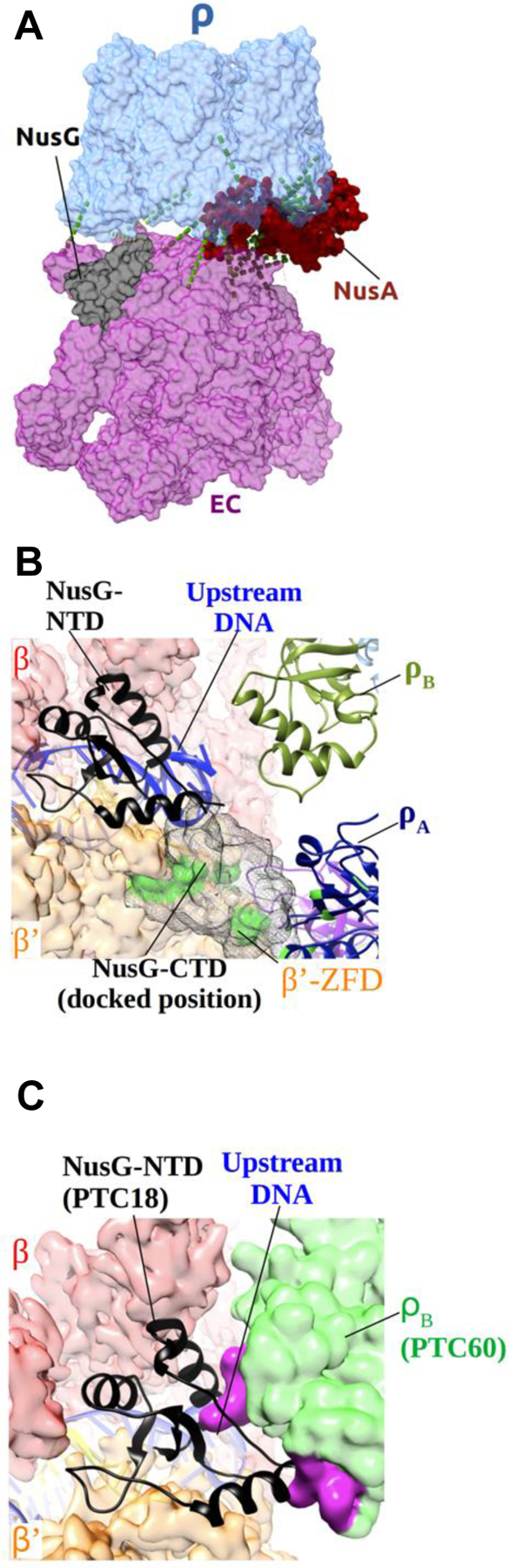
(A) Identified in-vitro crosslinks between PTC18 modules (see Table S4 for details). RNAP is colored purple; Rho is colored light blue; NusA is colored red; NusG is colored black. Green dashed lines represent the most abundant in-vitro crosslinks.
(B) Close-up view of NusG-NTD in PTC18. RNAP β and β’ subunits are shown as transparent red and orange surfaces, respectively. NusG-NTD is shown in black cartoon representation. ρB and ρA are shown as drab and navy ribbon diagrams, respectively. NusG-CTD, docked in a position consistent with in-vitro crosslinking sites (green spots), is shown as dark gray mesh surfaces.
(C) Close-up view of NusG-NTD and the upstream DNA in PTC18, also showing the modeled position of ρB of PTC60. The NusG-restrained upstream DNA is not compatible with the position of ρB in PTC60 as it would cause steric clashes. Rho subunit B in the position it adopts in PTC60 is shown as green transparent surface, and the residues that would cause steric clashes as solid magenta surface. The RNAP β subunit is shown as red transparent surface, the β’ subunit as orange transparent surface, NusG-NTD in black cartoon representation, and the upstream DNA in blue cartoon representation.
