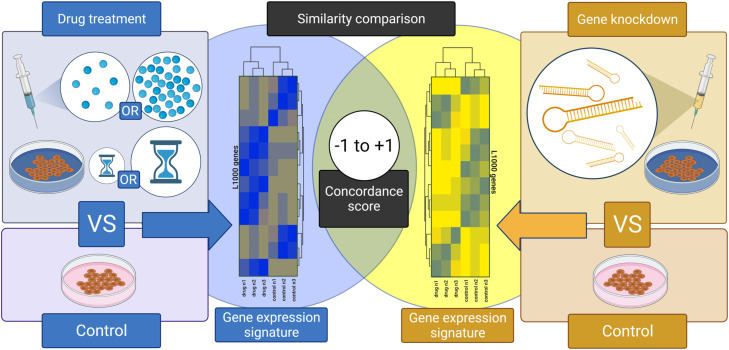
Fig. 1.
Experimental Design and Analytical Workflow. We generated differential gene expression signatures produced by fluoxetine, paroxetine, bupropion, or dexamethasone drug treatments across a variety of concentrations and treatment durations compared to control, untreated, wild-type cells. We also generated differential gene expression signatures produced by genetic knockdown of inflammatory genes related to the initiation and maintenance of IL6-mediated cytokine storm. Again, these signatures are in relation to control, untreated, wild-type cells. We then compared the drug treatment signatures with the genetic knockdown signatures to quantify similarity using concordance scores. When a drug treatment signature is highly concordant with a genetic knockdown signature, it generates a value approaching +1 and allows us to surmise that this drug and this genetic knockdown induce equivalent changes in gene expression. When no significant similarities exist, the concordance score approaches zero. When a drug treatment signature is highly discordant with a genetic knockdown signature, it generates a value approaching −1 and allows us to surmise that this drug and this genetic knockdown induce inverted changes in gene expression.