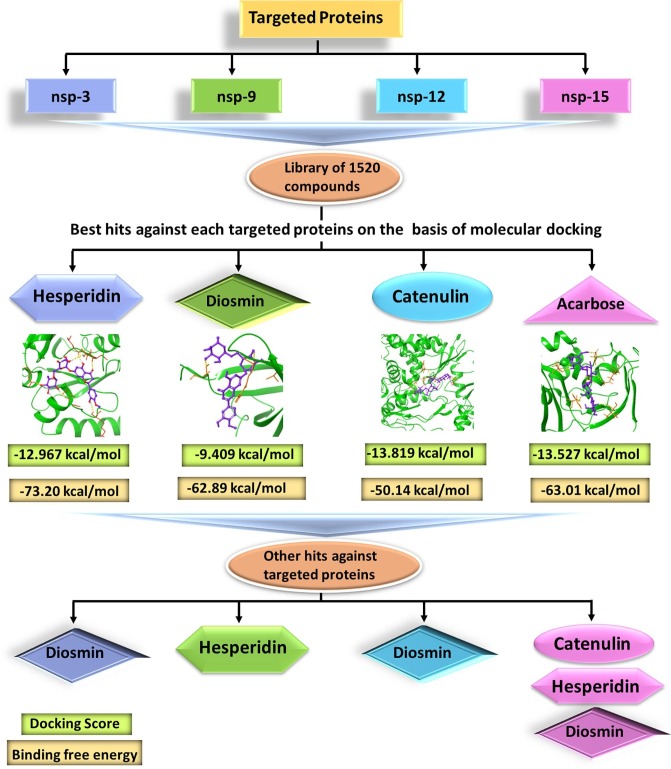
Fig. 1.
Flow chart of all the experiments carried out during the study. nsp3, nsp9, nsp12 and nsp15 protein of SARS-CoV-2 were screened with an approved library of 1520 compounds. The observation from the study revealed top compound against each targeted protein were: Hesperidin (1) (nsp3), Diosmin (6) (nsp9); Catenulin (13) (nsp12), and Acarbose (16) (nsp15) on the basis of molecular docking experiments. MD simulation was also carried out for these four compounds with respective protein, which revealed the stability of ligand–protein complex. Next, other hits compounds were also analyzed against each targeted protein and it was found that Diosmin was the one targeting all the proteins and hence considered for further studies.