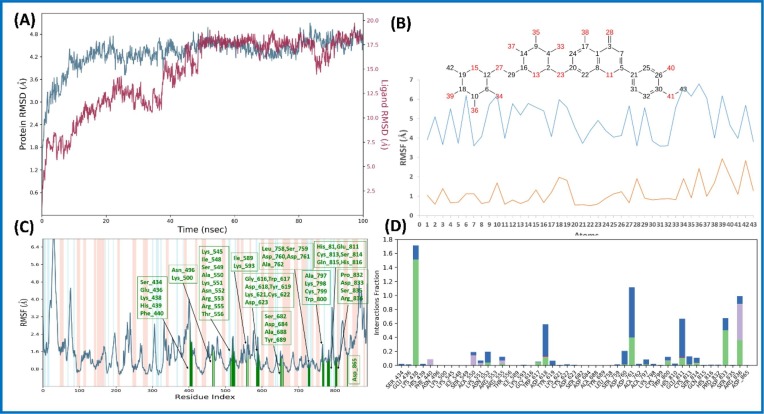
Fig. 11.
Molecular dynamic simulation analysis of Diosmin (6)-nsp12 complex: (A) RMSD plot; (B) ligand RMSF plot, where light blue line indicates the ligand fluctuations with respect to the binding site residues present on target protein and orange line shows the fluctuations where the ligand in each frame is aligned on the ligand in the first reference frame; (C) protein Cα RMSF plot, residues are shown in three letter code with green color belong to binding site interacting to Diosmin (6); and (D) ligand contacts histogram showing Diosmin (6)-nsp12 complex forming H-bond interaction (green), water interaction (blue), hydrophobic interaction (purple), and salt bridge interaction (pink) during a 100 ns simulation. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)