Fig. 4.
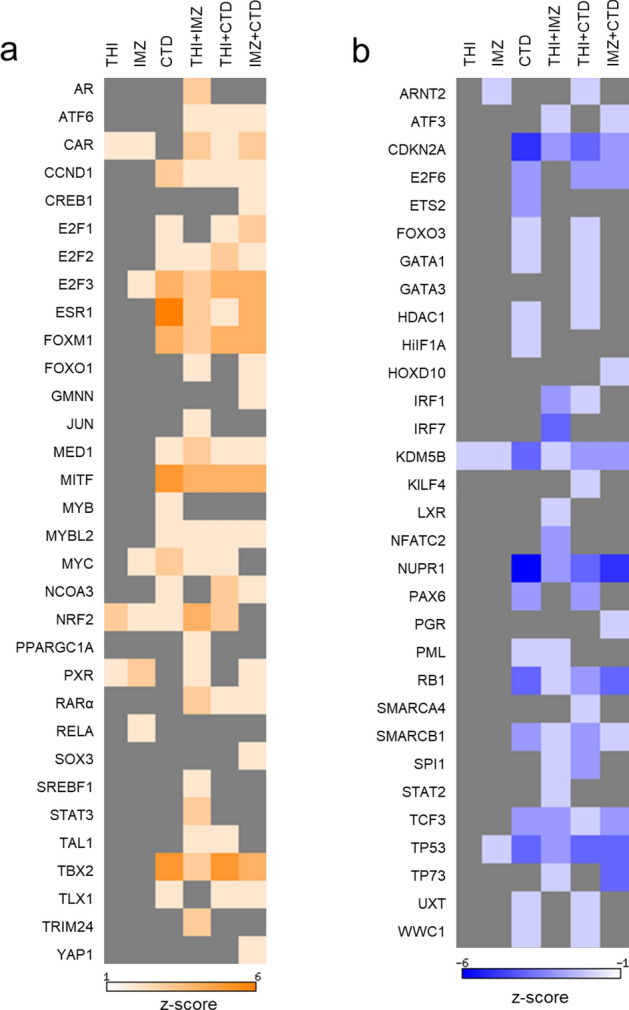
Transcription regulators predicted to be affected by THI, IMZ, CTD and their mixtures in rat livers. The list of genes significantly regulated by pesticides in rat hepatocytes as revealed by RNA-Seq (q < 0.05) was comparatively analyzed using Ingenuity Pathway Analysis. Upstream regulator analysis was performed and the p value (calculated by Fisher’s exact test right-tailed) was used to identify transcription regulators significantly associated with genes regulated by pesticides. The p value indicates the probability of association of a set of genes in the dataset with a transcription regulator by random chance alone. A p value < 0.05 was considered significant. The IPA activation z-score was used to predict transcription regulators that are increased or decreased by pesticides. Only transcription regulators with a z-score ≥ 2, predicting a significant increase (a), or a z-score ≤ − 2, predicting a significant decrease (b), were considered. Missing values (i.e., − 2 < z-score < 2) are depicted in gray
