Extended Data Fig. 6. Molecular Dynamics simulations reveal pleiotropic structural effects on UBs harboring lysine analogs.
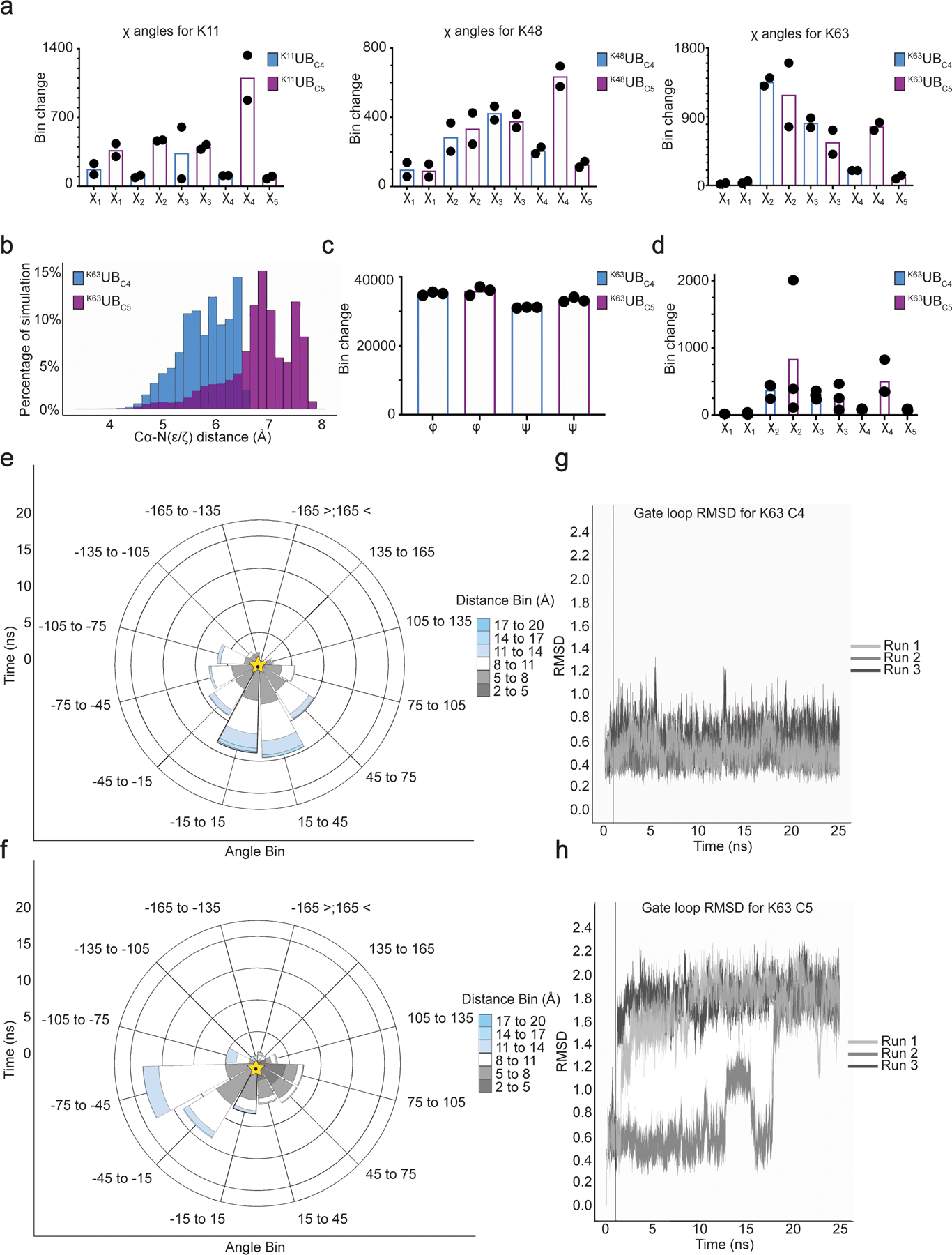
(a) Plot showing the degree of various side-chain rotamer interconversions for K11UBC5, K48UBC5, or K63UBC5 versus UBC4 acceptor UBs. Bins are divided by 120° intervals. (b) Distribution of the distances between lysine acceptor amine and Cα atoms for UBC4 versus UBC5 during 25 ns MD simulations (N=3 independent experiments) for the UBE2N~UB/UBE2V1/acceptor UB multi-subunit complex. Bins are divided by 10° intervals. (c) Plot showing the dynamics of phi and psi main-chain torsion angles for UBC4 or UBC5 acceptors in the UBE2N~UB/UBE2V1/acceptor UB multi-subunit complex. Bins are divided by 10° intervals. (d) Plot showing the dynamics of the side-chain rotamers for UBC4 or UBC5 acceptors in the UBE2N~UB/UBE2V1/acceptor UB multi-subunit complex. Bins are divided by 120° intervals. (e) Rose plot showing the distance and angle of the amine acceptor of UBC4 relative to the active-site during 25 ns MD simulations of the UBE2N~UB/UBE2V1/acceptor UB multi-subunit complex (N=3 independent experiments). Golden star indicated starting position. (f) same as (e), but with K63UBC5 (g) RMSD of gate loop during the trajectory for UBC4. (h) same as (g), except with K63UBC5.
