Extended Data Fig. 7. Di-UB chain formation reactions with UBC4 or UBC5 acceptors produce distinct results depending on the identity of the ubiquitin carrying enzyme.
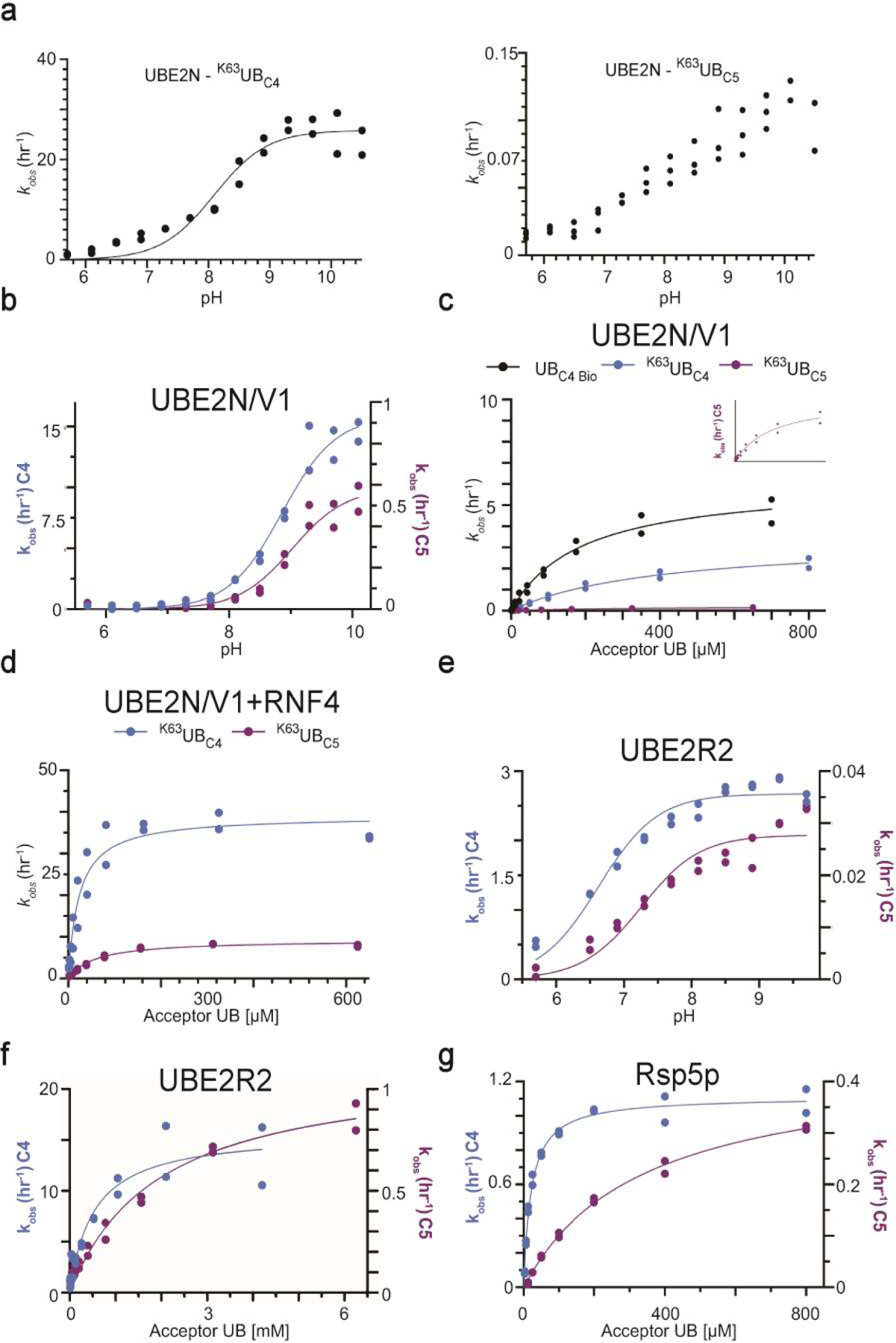
(a) Graph of the reactions velocities as a function of pH. performed in the presence of wild-type UBE2N/UBE2V1, radio-labeled K63R donor UB and either K63UBC4 or K63UBC5 acceptor UBs (b) same as (a), except with K92R UBE2N/UBE2V1. (c) Graph of the reaction velocities as a function of the acceptor UB concentration for UBE2N/UBE2V1. The inset shows the fit of the data to the model for reactions containing K63UBC5 acceptor UB since the magnitude of the velocities is far less than reactions containing UBC4. (d) same as (c) except in the presence of the RING domain of RNF4. (e) Graph of the reaction velocities performed as a function of pH, in the presence of UBE2R2 and radio-labeled K48R donor UB and either UBC4 or K48UBC5 acceptors. (f) Graph of the reaction velocities as a function of the acceptor UB concentration and their fit to the Michaelis-Menten model for UBE2R2. (g) same as (f), except in the presence of the yeast HECT E3 Rsp5p. N=2 independent experiments. For samples derived from the same experiment, gels were processed in parallel.
