Figure 4 |. The location of lysine analogs on acceptor UB impacts the distribution of diUB chain linkage types generated by the E2 enzyme UBE2D3.
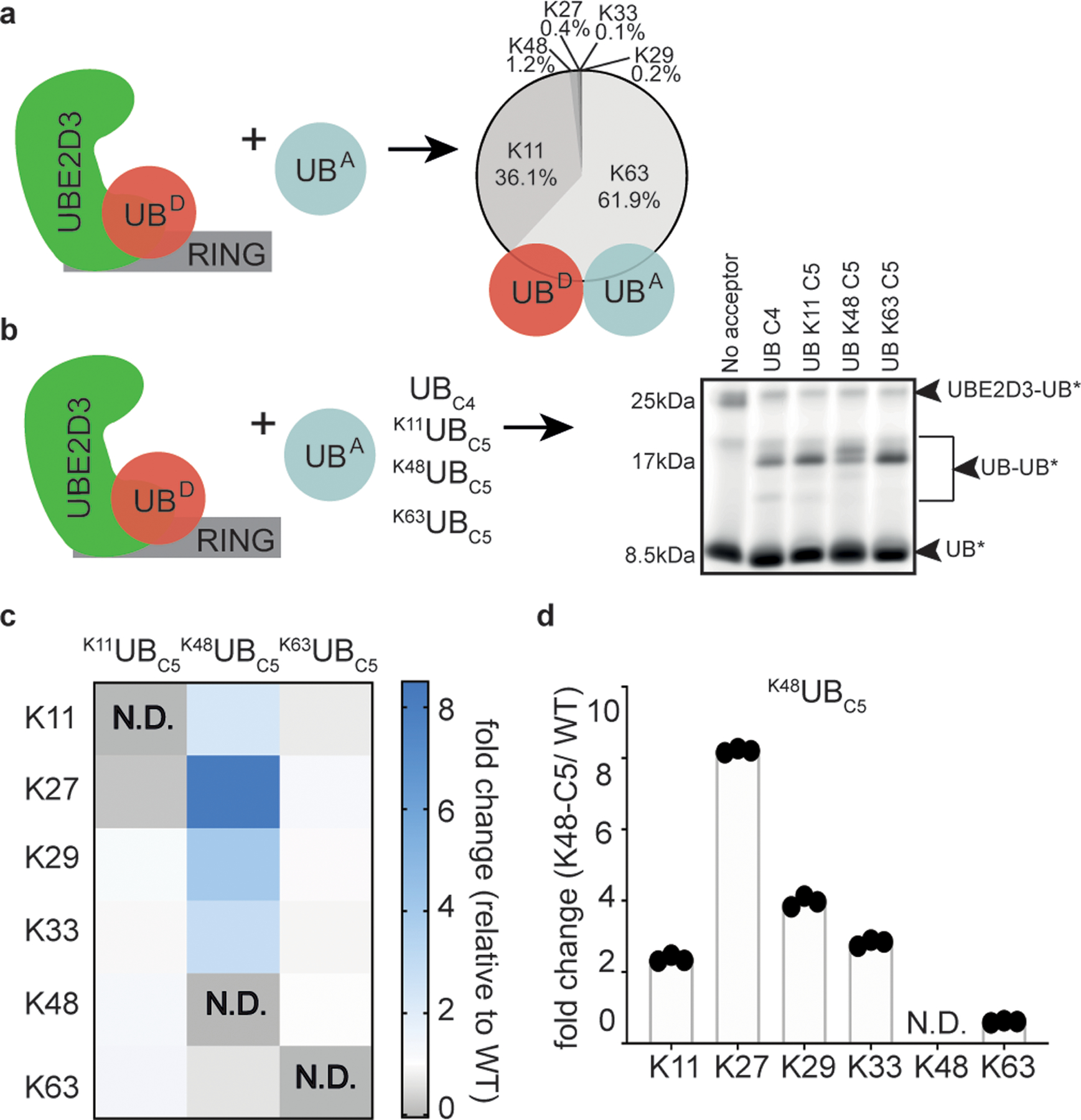
(a) Cartoon of experimental scheme, monitoring reactivity of UBE2D3~UB (D refers to the “donor” UB to be transferred from E2) in the presence of the E3 RING domain from RNF4 and formation of di-UB with UBC4 acceptor (UBA). The distribution of diUB linkage types generated is shown. (b) same as (a), except with fluorescent donor UB and various acceptor UBs (UBA). Notice that di-UB products with distinct electrophoretic mobilities were observed for each acceptor, indicating that the lysine analogs likely affect the di-UB chain linkage identity (N=2 independent experiments). (c) Relative fold changes of diUB linkage types for reactions with UBE2D3 and the E3 RING domain from RNF4 comparing K11UBC5, K48UBC5, or K63UBC5 acceptors with UBC4. N.D. is not-defined. (d) Plot showing the relative changes in UBE2D3/RNF4 generated di-UB chain linkages when comparing products containing K48UBC5 or UBC4 acceptors. For (c) and (d) N=3 technical replicates.
