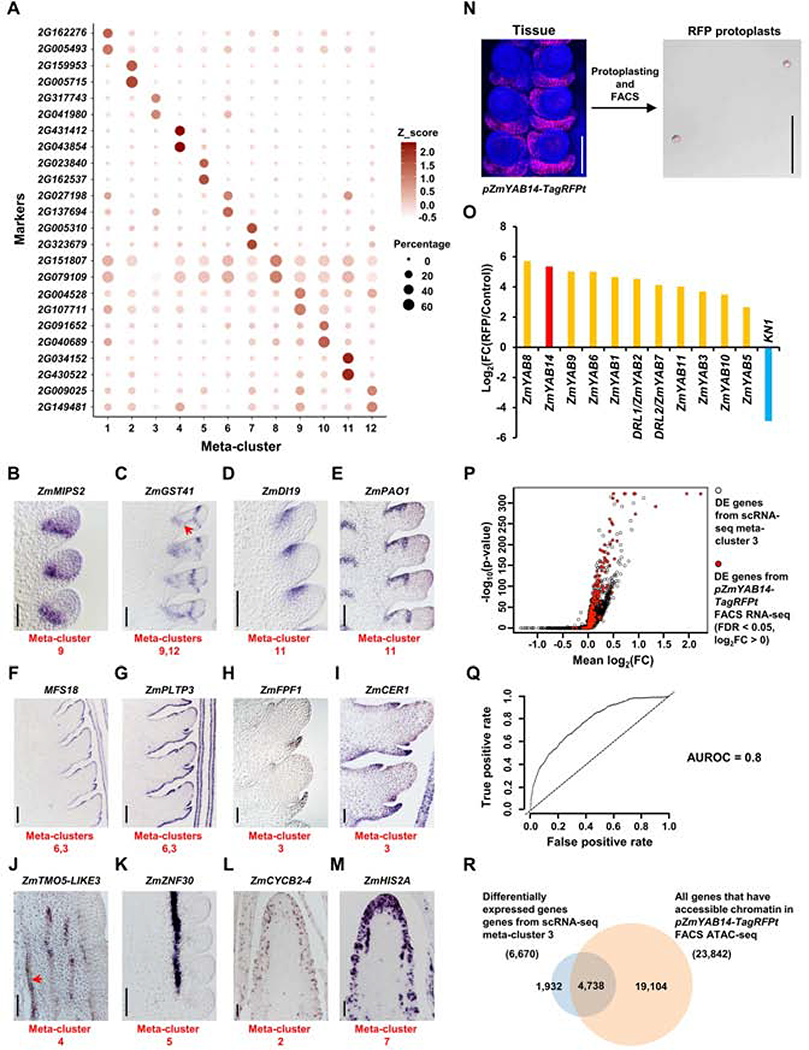
Figure 2: Validation of scRNA-seq by mRNA in situ and FACS RNA-seq.
(A) The top two marker genes of each meta-cluster are shown in dot plots with circle size indicating the percentage of cells expressing the marker and color representing Z_scored expression value. (B-M) mRNA in situ of meta-clusters marker genes validates the predicted identities: (B) ZmMIPS2, meristem boundary; (C) ZmGST41, meristem boundary (meta-cluster 9) and bundle sheath (meta-cluster 12, red arrow); (D-E) ZmDI19 (D) and ZmPAO1 (E), adaxial meristem periphery; (F-G) MFS18 (F) and ZmPLTP3 (G), meristem epidermis (meta-cluster 6) and determinate lateral organ (meta-cluster 3) epidermis; (H-I) ZmFPF1 (H) and ZmCER1 (I), determinate lateral organ; (J) ZmTMO5-LIKE3, xylem (red arrow indicates xylem vessels); (K) ZmZNF30, phloem. (L) ZmCYCB2–4, cell cycle G2/M phase; (M) ZmHIS2A, cell cycle S phase. Scale bar = 100μm. (N) Collection of RFP protoplasts from pZmYAB14-TagRFPt reporter line using FACS. Scale bar = 100μm. Three biological replicates are collected for FACS RNA-seq. One biological replicate is collected for FACS ATAC-seq. (O) Log2(fold change(FC)) of determinate lateral organ domain enriched markers, ZmYAB genes, and depleted marker, KN1, between RFP and total control protoplasts (Control) in FACS RNA-seq. (P) Volcano plot with 1-sided test positions the hits of enriched markers from pZmYAB14-TagRFPt FACS RNA-seq (red dots) on the ranked list of scRNA-seq differentially expressed (DE) genes from meta-cluster 3 (black circles). X-axis indicates the mean log2(FC) of DE genes between meta-cluster 3 and all other meta-clusters. Y-axis indicates corresponding −log10(p-value). (Q) pZmYAB14-TagRFPt FACS RNA-seq and scRNA-seq meta-cluster 3 have concordant differential gene expression patterns with Area Under the Receiver Operating Characteristics (AUROC) score = 0.8 (indicated by curved line; dashed line indicates the null (AUROC score = 0.5)). Axes indicates the true and false positive rate, the proportion of pZmYAB14-TagRFPt FACS RNA-seq enriched markers that do or do not match to scRNA-seq meta-cluster 3 enriched markers, respectively. (R) meta-cluster 3 DE genes are enriched in open chromatin in pZmYAB14-TagRFPt FACS sorted cells, see text for details.