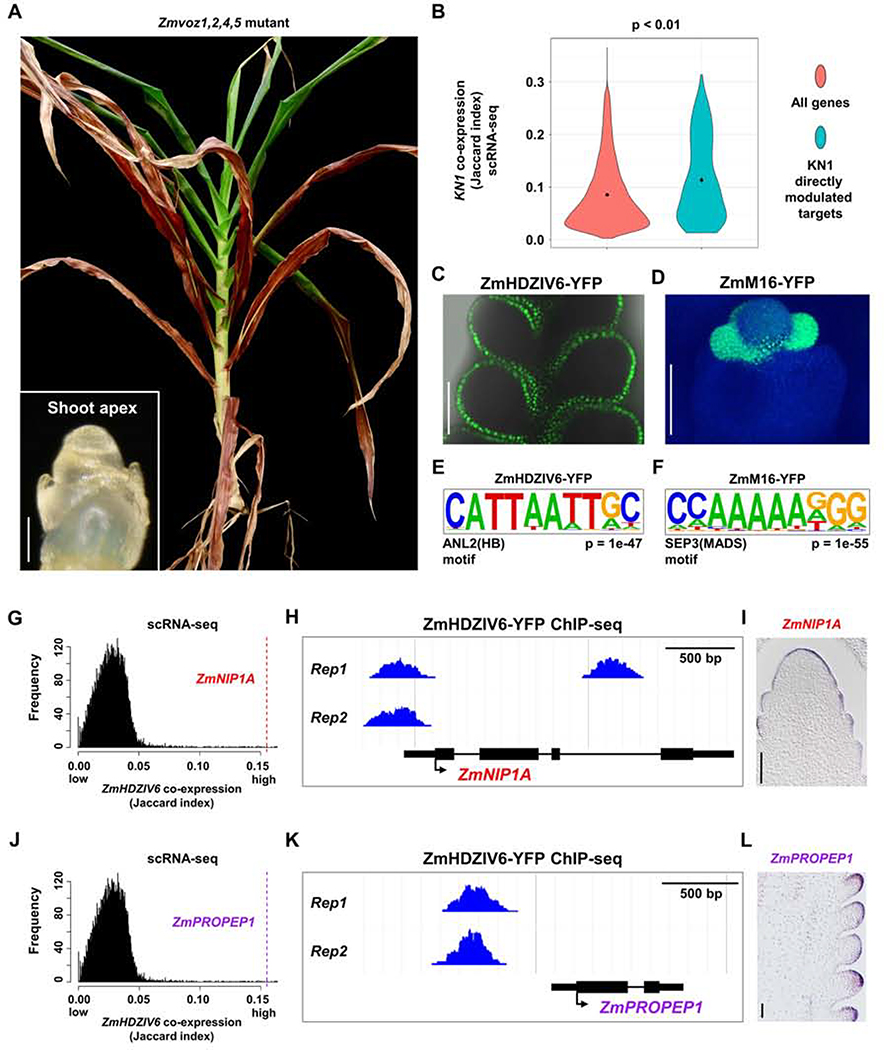
Figure 3: scRNA-seq Can Predict Genetic Redundancy and Aid in Predicting Transcriptional Regulatory Networks.
(A) Maize plant with CRISPR/cas9 knockout of four ZmVOZ paralogs fails to transition to flowering, as shown by two month old shoot apex (left bottom panel, scale bar = 100μm), and a 6 month old plant that lacks ears or tassel. (B) Directly modulated transcriptional targets of KN1 are significantly co-expressed with KN1 at the single-cell level; all maize genes are used as control (p < 0.01, one-way ANOVA with Tukey’s HSD). (C-D) Expression of TF translational fusion lines, ZmHDZIV6-YFP (C, merge of YFP channel and bright field) and ZmM16-YFP (D, merge of YFP and DAPI channels), used for two biological replicates of ChIP-seq. Scale bar = 100μm. (E-F) Expected motifs are significantly over-represented in bound peaks of ZmHDZIV6 (E) or ZmM16 (F). (G, J) ZmHDZIV6 candidate modulated targets, ZmNIP1A (G) and ZmPROPEP1 (J) are highly co-expressed with ZmHDZIV6 in scRNA-seq (Jaccard index = 0.155 for both targets). (H, K) ZmHDZIV6 bound peaks in ZmNIPA1 (H) and ZmPROPEP1 (K). (I, L) ZmNIP1A (I) and ZmPROPEP1 (L) are specifically expressed in the epidermis, by mRNA in situ. Scale bar = 100μm.