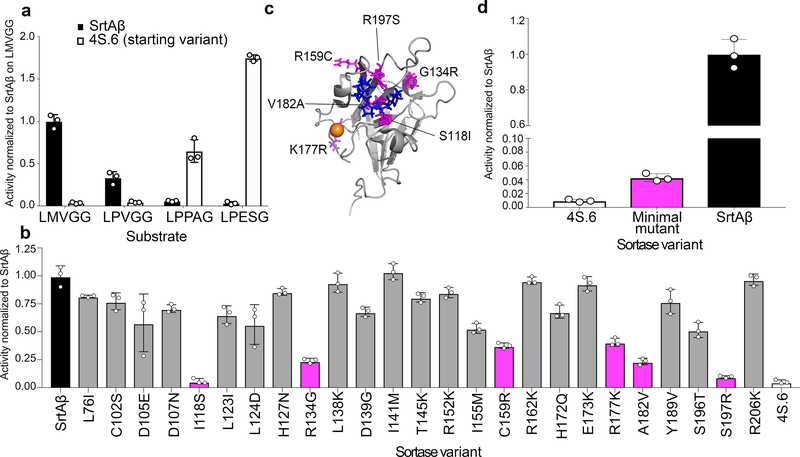
Figure 2. Activity profile and mutational analysis of SrtAβ.
(a) The evolved SrtAβ and the starting enzyme 4S.6 were displayed on yeast and assayed for their ability to catalyze transpeptidation on different substrates. (b) SrtAβ, 4S.6, and all 25 single-reversion mutants were displayed on yeast and assayed for their ability to catalyze transpeptidation between triglycine and Btn-LMVGG. Reversion mutants with activity less than half that of SrtAβ are highlighted in pink. (c) The predicted locations of the six reversion mutations that reduce SrtAβ activity by >50% are shown in pink on the NMR solution structure of wild-type S. aureus SrtA (PDB: 2KID). An LPXTG substrate analogue is blue, and the calcium ion required for activity is orange. Residues 118, 182, and 197 are part of the substrate binding pocket, while other residues are further from the active site. (d) The activity of 4S.6, a minimal mutant (4S.6 with S118I, G134R, R159C, K177R, V182A, and R197S mutations), and SrtAβ on Btn-LMVGG were compared by flow cytometry. Addition of these six mutations to 4S.6 improves activity on the LMVGG substrate, but is insufficient to confer the level of activity displayed by SrtAβ, highlighting the importance of other mutations. All graphs represent the mean of three replicates ± standard deviation. Activity is defined as the ratio of cell surface biotinylation (PE) to sortase expression level (FITC).