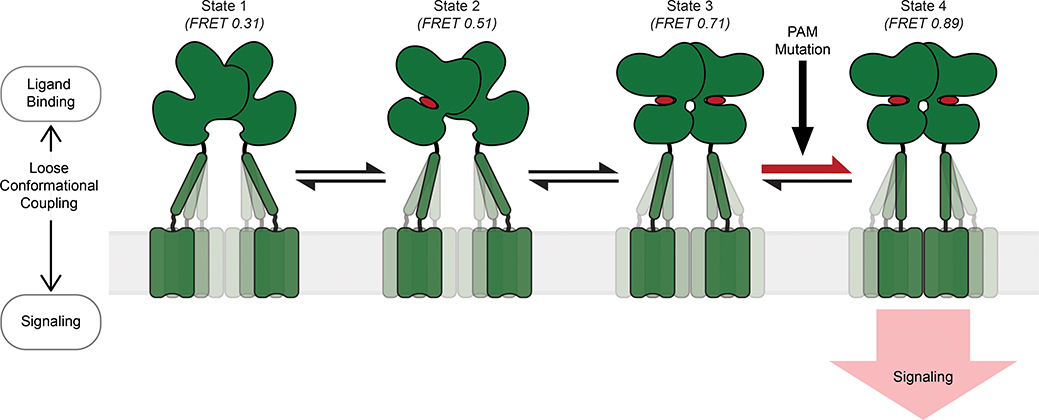
Fig. 5: Schematic of the stepwise activation model for mGluR2.
Four-state stepwise model for the mechanism of allosteric activation of mGluRs with receptors shown in green and ligand in red. Darker shading of CRDs and TMDs represent greater occupancy of a given conformation. State 1 (FRET=0.31) represents the most common CRD conformation of an unliganded receptor, where the CRDs are farthest apart. State 2 (FRET=0.51) represents the CRD conformation resulting from ligand binding to a single VFT domain. State 3 (FRET=0.71) represents the CRD conformation resulting from ligand binding to both VFT domains and prior to the formation of a stabilizing TMD interaction. State 4 (FRET=0.89) represents the CRD conformation resulting from ligand binding to both VFT domains and a stabilizing TMD interaction. State 4 is the active conformation of the receptor. PAM mutations can function by increasing transitions from state 3 to 4.