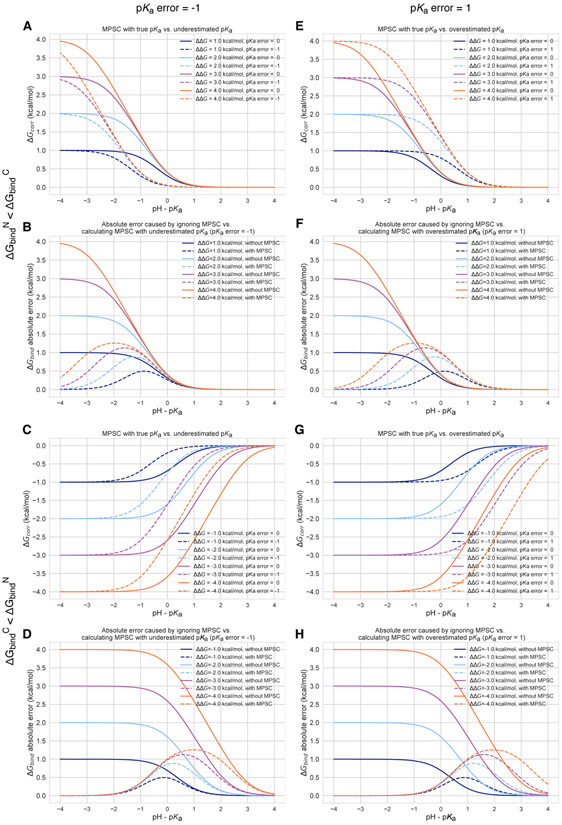
Figure 12. Inaccuracy of pKa prediction (± 1 unit) affects the the accuracy of MPSC and overall protein-ligand binding free energy calculations to varying degrees based on aqueous pKa and relative binding affinity of individual protonation states .
All calculations are made for 25°C, and a ligand with a single basic titratable group. A, C, E, and G show MPSC (ΔGcorr) calculated with true vs. inaccurate pKa. B, D, F, and H show the comparison of the absolute error to ΔGbind caused by ignoring the MPSC completely (solid lines) vs. calculating MPSC based in inaccurate pKa value (dashed lines). These plots provide guidance on when it is beneficial to include MPSC correction based on pKa error, pH - pKa, and ΔΔG.