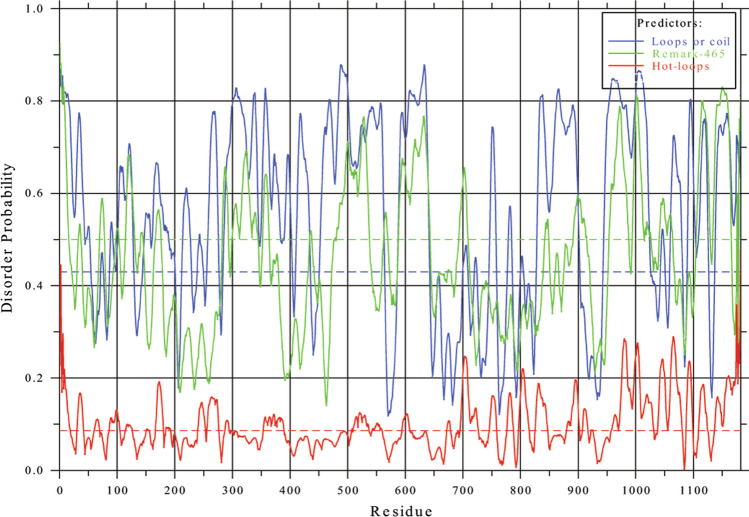
Fig. 1.
p140Cap and intrinsic disorder. Disorder probability of p140Cap human amino acid sequence (sequence ID: NP_079524.2), with DisEMBL computational tool. Figure downloaded from: DisEMBL https://dis.embl.de/, sequence ID: NP_079524, Intrinsic Protein Disorder Prediction 1.52). Predictions are shown according to each of the three different criteria to define the disorder level of protein residues, namely: loops or coils (blue line): loop assignment can be used as a necessary but not sufficient requirement for a disorder; -hot loops (red lines): constitute a subset of loops having a high degree of mobility as determined by C-alpha temperature (B-) factors. It follows that highly dynamic loops should be considered a protein disorder; missing coordinates in X-ray structure as defined by REMARK465 entries in PDB (green lines). Non-assigned electron densities in X-ray 3D structures most often reflect intrinsic disorder, and can be used in disorder predictions. For more detail, see [24]. The predicted probabilities are shown as curves along the sequences; scores should be compared to the corresponding random expectation value (dotted lines)